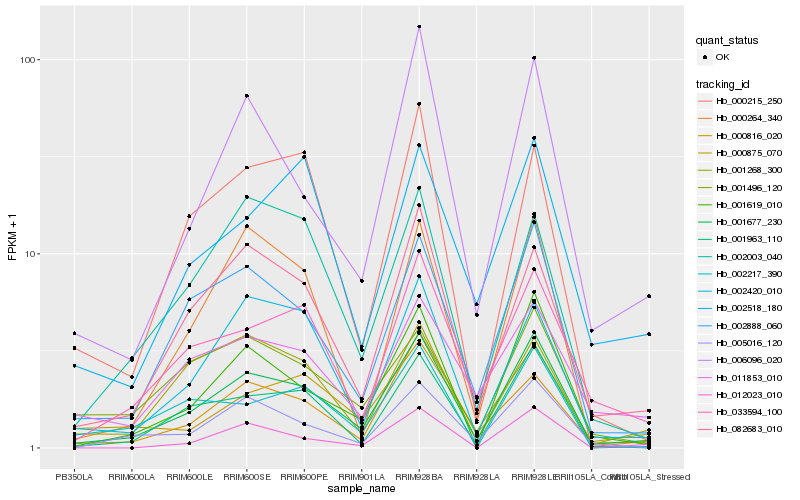
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001677_230 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002888_060 |
0.1113938965 |
- |
- |
PREDICTED: phospholipase SGR2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001619_010 |
0.1209812697 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET2a-like [Jatropha curcas] |
| 4 |
Hb_082683_010 |
0.1271134954 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_001963_110 |
0.1317886522 |
- |
- |
- |
| 6 |
Hb_006096_020 |
0.1380631086 |
- |
- |
Os06g0343500 [Oryza sativa Japonica Group] |
| 7 |
Hb_033594_100 |
0.1423315644 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable amino-acid acetyltransferase NAGS1, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_000816_020 |
0.144470697 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_002420_010 |
0.1480689561 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 10 |
Hb_005016_120 |
0.1498266704 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 11 |
Hb_000264_340 |
0.158476292 |
- |
- |
PREDICTED: callose synthase 11-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000875_070 |
0.1602381053 |
- |
- |
- |
| 13 |
Hb_000215_250 |
0.16252572 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 14 |
Hb_002217_390 |
0.1661556531 |
- |
- |
PREDICTED: galactokinase [Jatropha curcas] |
| 15 |
Hb_002003_040 |
0.1679880831 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 16 |
Hb_011853_010 |
0.1706875741 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 17 |
Hb_001268_300 |
0.170892998 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001496_120 |
0.170945189 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_002518_180 |
0.1711438164 |
- |
- |
hypothetical protein POPTR_0001s25460g [Populus trichocarpa] |
| 20 |
Hb_012023_010 |
0.1720114939 |
- |
- |
conserved hypothetical protein [Ricinus communis] |