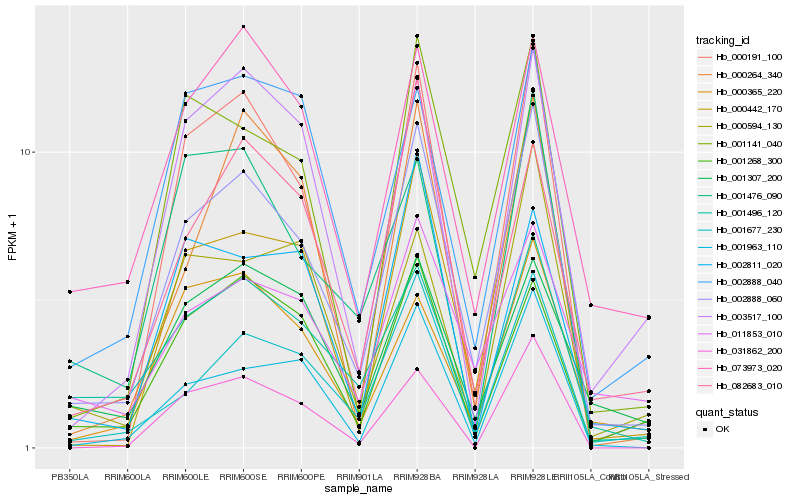
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002888_060 |
0.0 |
- |
- |
PREDICTED: phospholipase SGR2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001677_230 |
0.1113938965 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001141_040 |
0.120241491 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |
| 4 |
Hb_000191_100 |
0.1221587953 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 5 |
Hb_001496_120 |
0.1328144602 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 6 |
Hb_001963_110 |
0.133828026 |
- |
- |
- |
| 7 |
Hb_001268_300 |
0.137542101 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_082683_010 |
0.1378714532 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_000365_220 |
0.143183948 |
- |
- |
protein phosphatase-1, putative [Ricinus communis] |
| 10 |
Hb_000442_170 |
0.1443828295 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 11 |
Hb_002888_040 |
0.1502189304 |
- |
- |
hypothetical protein B456_009G267200 [Gossypium raimondii] |
| 12 |
Hb_011853_010 |
0.1516573751 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 13 |
Hb_003517_100 |
0.1517230671 |
transcription factor |
TF Family: GRAS |
- |
| 14 |
Hb_073973_020 |
0.1540510681 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 15 |
Hb_001476_090 |
0.154875579 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 3 [Jatropha curcas] |
| 16 |
Hb_000594_130 |
0.1549235356 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_001307_200 |
0.155769752 |
- |
- |
PREDICTED: uncharacterized protein LOC105644489 [Jatropha curcas] |
| 18 |
Hb_031862_200 |
0.1572038688 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At3g14460 [Vitis vinifera] |
| 19 |
Hb_000264_340 |
0.1582689882 |
- |
- |
PREDICTED: callose synthase 11-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_002811_020 |
0.1597259905 |
- |
- |
Cysteine synthase [Morus notabilis] |