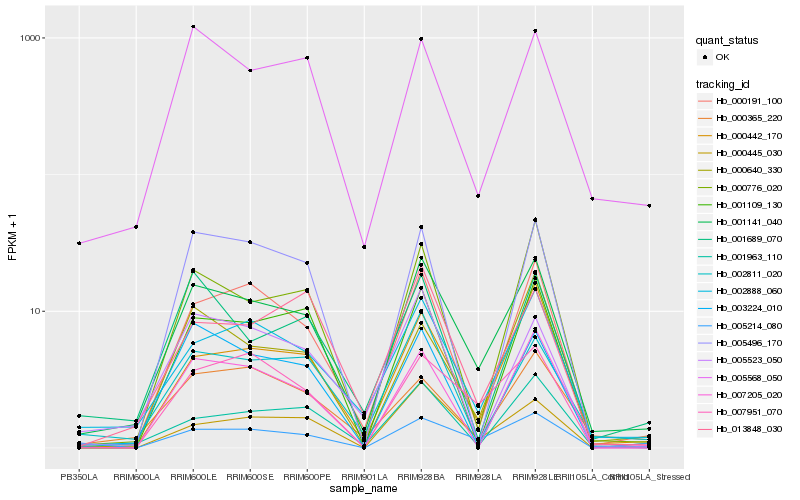
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001141_040 |
0.0 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |
| 2 |
Hb_002888_060 |
0.120241491 |
- |
- |
PREDICTED: phospholipase SGR2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000191_100 |
0.1384039788 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_005568_050 |
0.1466205381 |
- |
- |
ascorbate peroxidase [Hevea brasiliensis] |
| 5 |
Hb_002811_020 |
0.1476509666 |
- |
- |
Cysteine synthase [Morus notabilis] |
| 6 |
Hb_000442_170 |
0.1511388664 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 7 |
Hb_000776_020 |
0.1544373494 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_25594 [Jatropha curcas] |
| 8 |
Hb_007951_070 |
0.1547547449 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 9 |
Hb_001963_110 |
0.1613865158 |
- |
- |
- |
| 10 |
Hb_000445_030 |
0.166613496 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase 5 [Jatropha curcas] |
| 11 |
Hb_007205_020 |
0.1674180129 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 12 |
Hb_003224_010 |
0.1688343456 |
- |
- |
- |
| 13 |
Hb_000365_220 |
0.1693550259 |
- |
- |
protein phosphatase-1, putative [Ricinus communis] |
| 14 |
Hb_013848_030 |
0.169543619 |
- |
- |
PREDICTED: probable aminotransferase TAT2 [Jatropha curcas] |
| 15 |
Hb_005523_050 |
0.1702218484 |
- |
- |
hypothetical protein JCGZ_09175 [Jatropha curcas] |
| 16 |
Hb_005214_080 |
0.1702880044 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005496_170 |
0.1720170786 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 18 |
Hb_001689_070 |
0.1728179292 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_000640_330 |
0.1729405425 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001109_130 |
0.1748810051 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 136 isoform X1 [Jatropha curcas] |