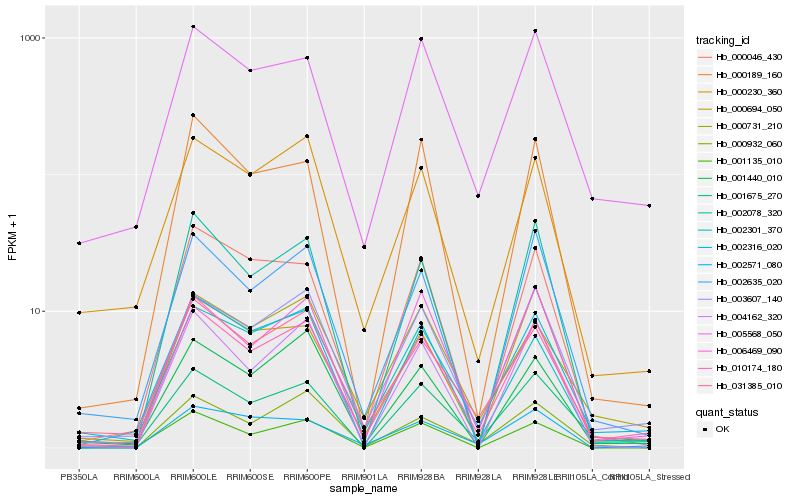
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010174_180 |
0.0 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_002316_020 |
0.07529422 |
- |
- |
PREDICTED: uncharacterized protein LOC105640407 [Jatropha curcas] |
| 3 |
Hb_003607_140 |
0.0756157517 |
- |
- |
3-ketoacyl-CoA thiolase B, putative [Ricinus communis] |
| 4 |
Hb_031385_010 |
0.0973790181 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 5 |
Hb_002635_020 |
0.1058584935 |
- |
- |
hypothetical protein POPTR_0018s09790g [Populus trichocarpa] |
| 6 |
Hb_001675_270 |
0.1072727233 |
- |
- |
PREDICTED: probable inactive nicotinamidase At3g16190 [Jatropha curcas] |
| 7 |
Hb_000731_210 |
0.1089818891 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006469_090 |
0.1096652383 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: myosin-11 [Jatropha curcas] |
| 9 |
Hb_001440_010 |
0.1110860709 |
- |
- |
PREDICTED: armadillo repeat-containing kinesin-like protein 1 [Populus euphratica] |
| 10 |
Hb_002301_370 |
0.1131693884 |
- |
- |
hypothetical protein JCGZ_21454 [Jatropha curcas] |
| 11 |
Hb_002078_320 |
0.1138258402 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1 [Jatropha curcas] |
| 12 |
Hb_004162_320 |
0.1139087743 |
- |
- |
PREDICTED: uncharacterized protein LOC105628009 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000230_360 |
0.1140105845 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 14 |
Hb_005568_050 |
0.1160808065 |
- |
- |
ascorbate peroxidase [Hevea brasiliensis] |
| 15 |
Hb_000694_050 |
0.1175260706 |
- |
- |
Nuclease PA3, putative [Ricinus communis] |
| 16 |
Hb_000189_160 |
0.1188756205 |
- |
- |
PREDICTED: GDP-mannose 3,5-epimerase 2 [Eucalyptus grandis] |
| 17 |
Hb_001135_010 |
0.1193283538 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 18 |
Hb_000046_430 |
0.1195246119 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 19 |
Hb_002571_080 |
0.1201129263 |
- |
- |
secondary cell wall-related glycosyltransferase family 14 [Populus tremula x Populus tremuloides] |
| 20 |
Hb_000932_060 |
0.12050839 |
- |
- |
conserved hypothetical protein [Ricinus communis] |