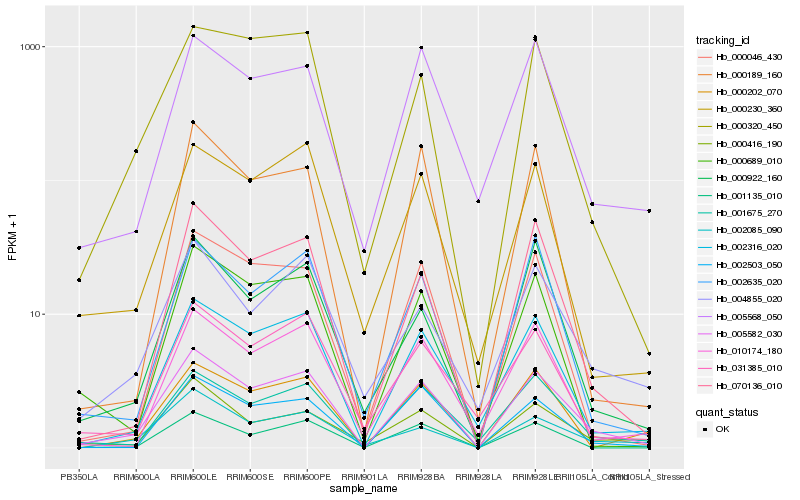
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005582_030 |
0.0 |
- |
- |
PREDICTED: protein N-lysine methyltransferase METTL21A-like [Jatropha curcas] |
| 2 |
Hb_002316_020 |
0.1135930774 |
- |
- |
PREDICTED: uncharacterized protein LOC105640407 [Jatropha curcas] |
| 3 |
Hb_001675_270 |
0.121977906 |
- |
- |
PREDICTED: probable inactive nicotinamidase At3g16190 [Jatropha curcas] |
| 4 |
Hb_070136_010 |
0.1303929471 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 5 |
Hb_002503_050 |
0.1323621405 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 6 |
Hb_000320_450 |
0.1396307501 |
- |
- |
secretory peroxidase [Hevea brasiliensis] |
| 7 |
Hb_000230_360 |
0.1418715668 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 8 |
Hb_002635_020 |
0.1423242741 |
- |
- |
hypothetical protein POPTR_0018s09790g [Populus trichocarpa] |
| 9 |
Hb_001135_010 |
0.145413392 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 10 |
Hb_010174_180 |
0.145485197 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_000046_430 |
0.1463639177 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 12 |
Hb_002085_090 |
0.1472192947 |
- |
- |
- |
| 13 |
Hb_000689_010 |
0.1474295346 |
desease resistance |
Gene Name: LRR_8 |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 14 |
Hb_000416_190 |
0.1485255632 |
- |
- |
- |
| 15 |
Hb_005568_050 |
0.1488596303 |
- |
- |
ascorbate peroxidase [Hevea brasiliensis] |
| 16 |
Hb_004855_020 |
0.1493396485 |
- |
- |
hypothetical protein POPTR_0006s10010g [Populus trichocarpa] |
| 17 |
Hb_000922_160 |
0.149787132 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
putative 1-deoxy-D-xylulose 5-phosphate reductoisomerase [Hevea brasiliensis] |
| 18 |
Hb_000189_160 |
0.150224171 |
- |
- |
PREDICTED: GDP-mannose 3,5-epimerase 2 [Eucalyptus grandis] |
| 19 |
Hb_000202_070 |
0.1506481936 |
- |
- |
putative RNA-directed DNA polymerase (Reverse transcriptase) [Malus domestica] |
| 20 |
Hb_031385_010 |
0.1509263832 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |