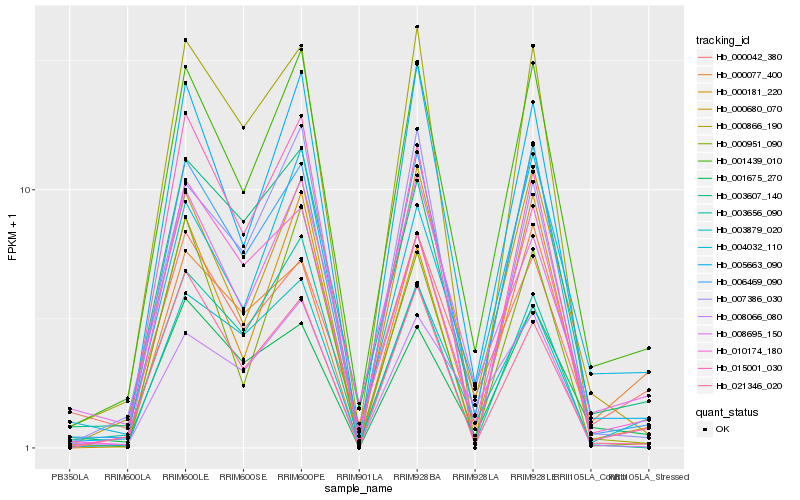
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001439_010 |
0.0 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 5-like [Jatropha curcas] |
| 2 |
Hb_006469_090 |
0.0770281172 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: myosin-11 [Jatropha curcas] |
| 3 |
Hb_000680_070 |
0.0831478638 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005663_090 |
0.0915753405 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 7 [Jatropha curcas] |
| 5 |
Hb_003607_140 |
0.096231105 |
- |
- |
3-ketoacyl-CoA thiolase B, putative [Ricinus communis] |
| 6 |
Hb_003879_020 |
0.1047195838 |
- |
- |
PREDICTED: MATE efflux family protein 1-like [Jatropha curcas] |
| 7 |
Hb_004032_110 |
0.1127285739 |
- |
- |
PREDICTED: putative methylesterase 14, chloroplastic [Jatropha curcas] |
| 8 |
Hb_008695_150 |
0.1165910875 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 7 isoform X1 [Jatropha curcas] |
| 9 |
Hb_010174_180 |
0.1213423082 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_001675_270 |
0.1235699648 |
- |
- |
PREDICTED: probable inactive nicotinamidase At3g16190 [Jatropha curcas] |
| 11 |
Hb_000181_220 |
0.1268394517 |
- |
- |
PREDICTED: MATE efflux family protein LAL5-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_000077_400 |
0.1274970369 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 13 |
Hb_008066_080 |
0.1301370119 |
- |
- |
PREDICTED: GDSL esterase/lipase 1-like [Jatropha curcas] |
| 14 |
Hb_021346_020 |
0.1303202457 |
- |
- |
hypothetical protein JCGZ_26746 [Jatropha curcas] |
| 15 |
Hb_000866_190 |
0.1314828347 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_000042_380 |
0.1325889752 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 [Jatropha curcas] |
| 17 |
Hb_003656_090 |
0.1359261806 |
- |
- |
PREDICTED: protein Brevis radix-like 4 [Jatropha curcas] |
| 18 |
Hb_015001_030 |
0.1365569919 |
- |
- |
inosine-uridine preferring nucleoside hydrolase family protein [Populus trichocarpa] |
| 19 |
Hb_007386_030 |
0.137152508 |
- |
- |
PREDICTED: formin-like protein 2 [Jatropha curcas] |
| 20 |
Hb_000951_090 |
0.1373871585 |
- |
- |
Oligopeptide transporter, putative [Ricinus communis] |