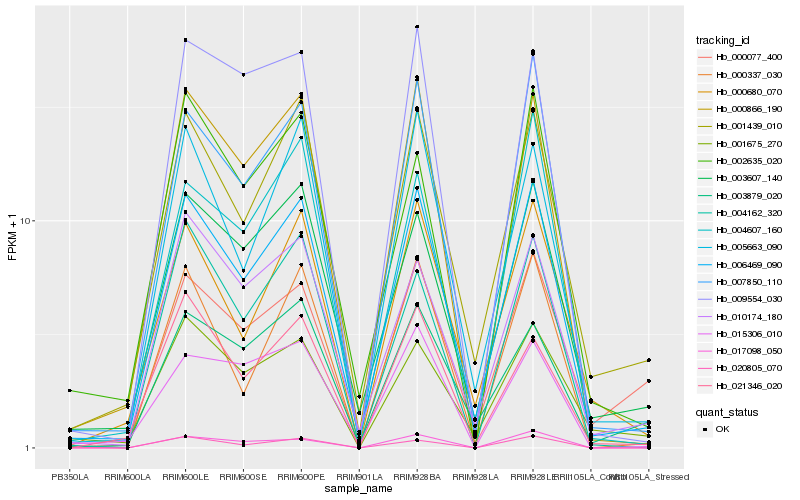
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006469_090 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: myosin-11 [Jatropha curcas] |
| 2 |
Hb_001439_010 |
0.0770281172 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 5-like [Jatropha curcas] |
| 3 |
Hb_007850_110 |
0.0828598476 |
- |
- |
MADS-box transcription factor 3 [Hevea brasiliensis] |
| 4 |
Hb_000866_190 |
0.0874652668 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_003607_140 |
0.0947962222 |
- |
- |
3-ketoacyl-CoA thiolase B, putative [Ricinus communis] |
| 6 |
Hb_005663_090 |
0.1012781979 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 7 [Jatropha curcas] |
| 7 |
Hb_000680_070 |
0.1021622416 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_010174_180 |
0.1096652383 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 9 |
Hb_000337_030 |
0.1152570988 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 10 |
Hb_020805_070 |
0.1204367769 |
- |
- |
PREDICTED: MLO protein homolog 1-like [Jatropha curcas] |
| 11 |
Hb_004607_160 |
0.1217100175 |
- |
- |
PREDICTED: uncharacterized protein LOC105134931 [Populus euphratica] |
| 12 |
Hb_003879_020 |
0.1246941414 |
- |
- |
PREDICTED: MATE efflux family protein 1-like [Jatropha curcas] |
| 13 |
Hb_017098_050 |
0.1258573401 |
- |
- |
hypothetical protein POPTR_0001s03540g [Populus trichocarpa] |
| 14 |
Hb_001675_270 |
0.1261797356 |
- |
- |
PREDICTED: probable inactive nicotinamidase At3g16190 [Jatropha curcas] |
| 15 |
Hb_002635_020 |
0.1275825193 |
- |
- |
hypothetical protein POPTR_0018s09790g [Populus trichocarpa] |
| 16 |
Hb_000077_400 |
0.1278397618 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_004162_320 |
0.1279880258 |
- |
- |
PREDICTED: uncharacterized protein LOC105628009 isoform X1 [Jatropha curcas] |
| 18 |
Hb_009554_030 |
0.128393517 |
- |
- |
PREDICTED: chlorophyll synthase, chloroplastic [Prunus mume] |
| 19 |
Hb_021346_020 |
0.1290342404 |
- |
- |
hypothetical protein JCGZ_26746 [Jatropha curcas] |
| 20 |
Hb_015306_010 |
0.130614876 |
- |
- |
PREDICTED: uncharacterized protein LOC105636357 [Jatropha curcas] |