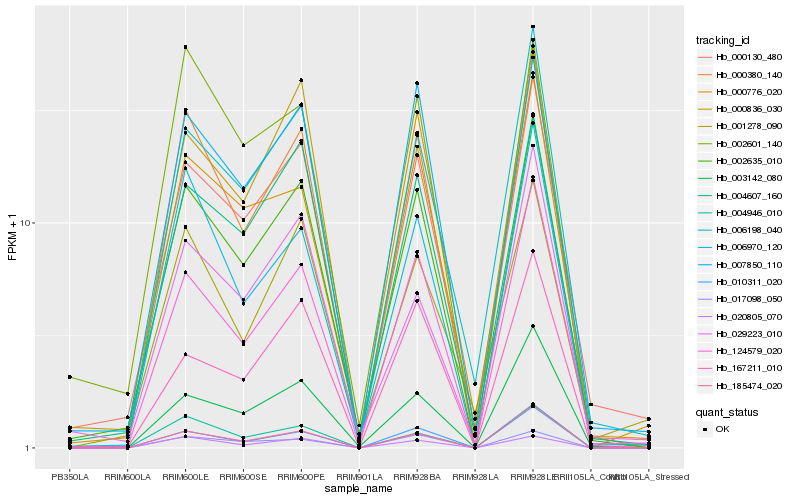
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002635_010 |
0.0 |
- |
- |
homogentisate phytyl transferase [Hevea brasiliensis] |
| 2 |
Hb_000130_480 |
0.0683800642 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase SETD1B [Jatropha curcas] |
| 3 |
Hb_000380_140 |
0.0722536836 |
- |
- |
PREDICTED: uncharacterized protein LOC105628069 [Jatropha curcas] |
| 4 |
Hb_004607_160 |
0.0820229686 |
- |
- |
PREDICTED: uncharacterized protein LOC105134931 [Populus euphratica] |
| 5 |
Hb_124579_020 |
0.0865748562 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 6 |
Hb_007850_110 |
0.0904511284 |
- |
- |
MADS-box transcription factor 3 [Hevea brasiliensis] |
| 7 |
Hb_003142_080 |
0.0923275548 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 8 |
Hb_010311_020 |
0.0923520823 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 9 |
Hb_006198_040 |
0.0948877542 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000836_030 |
0.0997073068 |
- |
- |
ubiquitin-conjugating enzyme h, putative [Ricinus communis] |
| 11 |
Hb_185474_020 |
0.1003939748 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 12 |
Hb_006970_120 |
0.1106317641 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_167211_010 |
0.1171298588 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_004946_010 |
0.1184242446 |
- |
- |
hypothetical protein POPTR_0013s035502g, partial [Populus trichocarpa] |
| 15 |
Hb_001278_090 |
0.1215082344 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 16 |
Hb_002601_140 |
0.1219909964 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 17 |
Hb_017098_050 |
0.1221784034 |
- |
- |
hypothetical protein POPTR_0001s03540g [Populus trichocarpa] |
| 18 |
Hb_020805_070 |
0.1270924641 |
- |
- |
PREDICTED: MLO protein homolog 1-like [Jatropha curcas] |
| 19 |
Hb_000776_020 |
0.1273014738 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_25594 [Jatropha curcas] |
| 20 |
Hb_029223_010 |
0.1278167335 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |