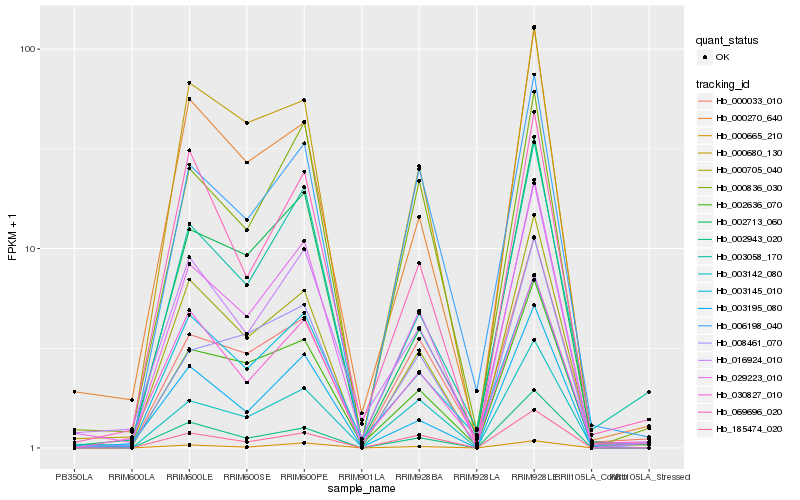
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029223_010 |
0.0 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |
| 2 |
Hb_016924_010 |
0.0711717214 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002636_070 |
0.0812452345 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 4 |
Hb_000705_040 |
0.0879831155 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003142_080 |
0.0935383172 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 6 |
Hb_000270_640 |
0.0995542048 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 7 |
Hb_000836_030 |
0.0998107246 |
- |
- |
ubiquitin-conjugating enzyme h, putative [Ricinus communis] |
| 8 |
Hb_002713_060 |
0.1019444545 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.4 [Jatropha curcas] |
| 9 |
Hb_003145_010 |
0.1035338526 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 10 |
Hb_000033_010 |
0.1036904015 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 11 |
Hb_003058_170 |
0.1044063634 |
- |
- |
PREDICTED: F-box protein At2g26850 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003195_080 |
0.105316529 |
- |
- |
PREDICTED: cytochrome P450 87A3-like [Jatropha curcas] |
| 13 |
Hb_006198_040 |
0.1056428663 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_008461_070 |
0.1129489706 |
- |
- |
PREDICTED: ethylene-overproduction protein 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000665_210 |
0.1139585551 |
- |
- |
PREDICTED: synaptotagmin-4 [Jatropha curcas] |
| 16 |
Hb_185474_020 |
0.11554845 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 17 |
Hb_030827_010 |
0.1158592828 |
transcription factor |
TF Family: HB |
Homeobox-leucine zipper ROC6 [Gossypium arboreum] |
| 18 |
Hb_000680_130 |
0.1163279876 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 19 |
Hb_002943_020 |
0.1184639501 |
- |
- |
hypothetical protein POPTR_0075s00240g, partial [Populus trichocarpa] |
| 20 |
Hb_069696_020 |
0.1192563471 |
- |
- |
PREDICTED: protochlorophyllide-dependent translocon component 52, chloroplastic-like isoform X2 [Jatropha curcas] |