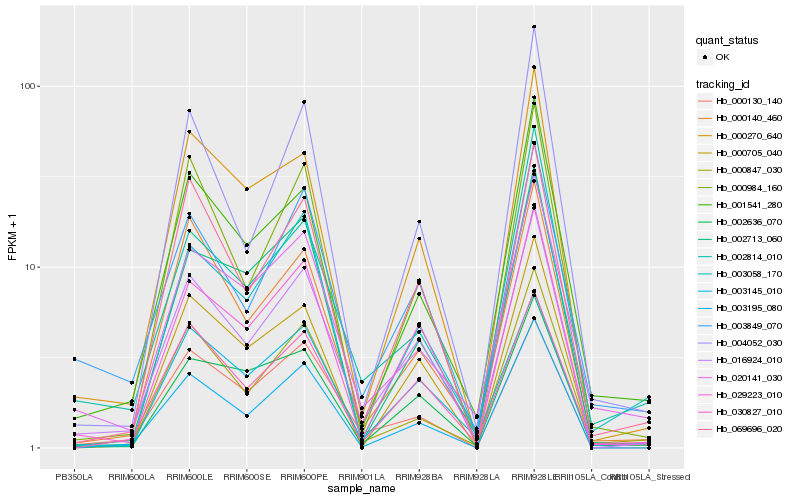
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_016924_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_029223_010 |
0.0711717214 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |
| 3 |
Hb_003195_080 |
0.095104714 |
- |
- |
PREDICTED: cytochrome P450 87A3-like [Jatropha curcas] |
| 4 |
Hb_000270_640 |
0.0975739736 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 5 |
Hb_003145_010 |
0.1036857038 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 6 |
Hb_030827_010 |
0.1038711316 |
transcription factor |
TF Family: HB |
Homeobox-leucine zipper ROC6 [Gossypium arboreum] |
| 7 |
Hb_069696_020 |
0.1071436465 |
- |
- |
PREDICTED: protochlorophyllide-dependent translocon component 52, chloroplastic-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_003849_070 |
0.1075903853 |
- |
- |
PREDICTED: uncharacterized protein LOC105644690 [Jatropha curcas] |
| 9 |
Hb_000705_040 |
0.1124697216 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001541_280 |
0.1125387082 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 11 |
Hb_002636_070 |
0.1128709799 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 12 |
Hb_000140_460 |
0.113631534 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 13 |
Hb_000984_160 |
0.1143189396 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000847_030 |
0.1159373996 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 2 [Jatropha curcas] |
| 15 |
Hb_020141_030 |
0.1179157269 |
- |
- |
solanesyl diphosphate synthase, putative [Ricinus communis] |
| 16 |
Hb_000130_140 |
0.1191245622 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105639203 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003058_170 |
0.1205247377 |
- |
- |
PREDICTED: F-box protein At2g26850 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004052_030 |
0.1222503372 |
- |
- |
PREDICTED: uncharacterized protein LOC105640637 [Jatropha curcas] |
| 19 |
Hb_002713_060 |
0.1237256721 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.4 [Jatropha curcas] |
| 20 |
Hb_002814_010 |
0.1251116201 |
- |
- |
PREDICTED: adenylate kinase 5, chloroplastic [Jatropha curcas] |