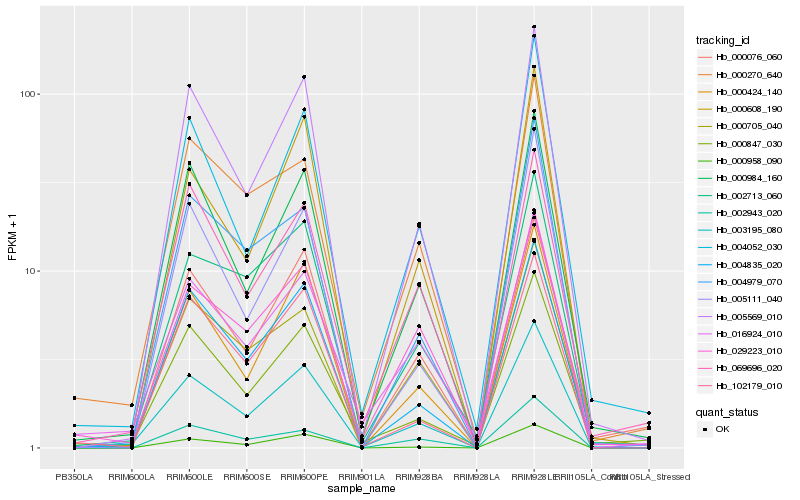
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003195_080 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 87A3-like [Jatropha curcas] |
| 2 |
Hb_005569_010 |
0.0716037348 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 3 |
Hb_004835_020 |
0.0841851573 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 4 |
Hb_000984_160 |
0.0848899035 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000424_140 |
0.0883977227 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_016924_010 |
0.095104714 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004052_030 |
0.0975060002 |
- |
- |
PREDICTED: uncharacterized protein LOC105640637 [Jatropha curcas] |
| 8 |
Hb_000608_190 |
0.0975737196 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
PREDICTED: isopentenyl-diphosphate Delta-isomerase I [Jatropha curcas] |
| 9 |
Hb_000847_030 |
0.0982129649 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 2 [Jatropha curcas] |
| 10 |
Hb_002713_060 |
0.100334717 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.4 [Jatropha curcas] |
| 11 |
Hb_000958_090 |
0.1016631113 |
- |
- |
PREDICTED: cation/H(+) antiporter 28 [Jatropha curcas] |
| 12 |
Hb_000705_040 |
0.1037677565 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_029223_010 |
0.105316529 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |
| 14 |
Hb_004979_070 |
0.1067061693 |
- |
- |
wall-associated receptor kinase galacturonan-binding protein [Medicago truncatula] |
| 15 |
Hb_000270_640 |
0.108856589 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 16 |
Hb_000076_060 |
0.111838607 |
- |
- |
hypothetical protein RCOM_0586940 [Ricinus communis] |
| 17 |
Hb_002943_020 |
0.1129646439 |
- |
- |
hypothetical protein POPTR_0075s00240g, partial [Populus trichocarpa] |
| 18 |
Hb_005111_040 |
0.1130645858 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 19 |
Hb_069696_020 |
0.1134843278 |
- |
- |
PREDICTED: protochlorophyllide-dependent translocon component 52, chloroplastic-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_102179_010 |
0.113761182 |
- |
- |
protein binding protein, putative [Ricinus communis] |