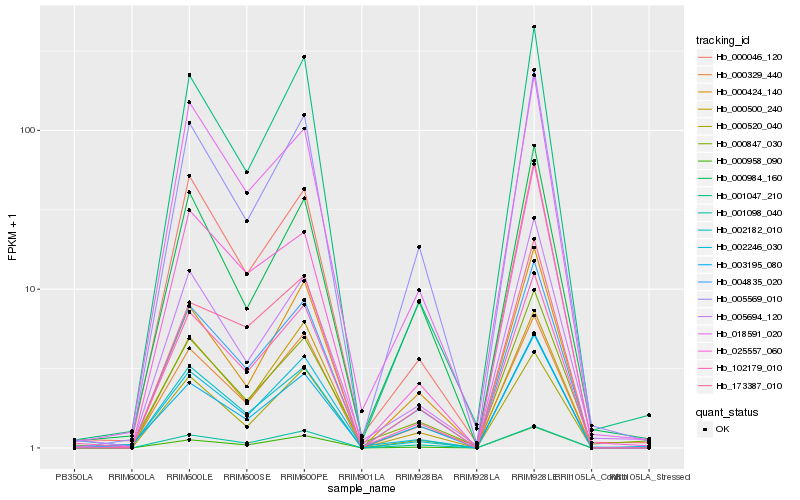
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004835_020 |
0.0 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 2 |
Hb_102179_010 |
0.0441464182 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_005569_010 |
0.0469109468 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 4 |
Hb_000329_440 |
0.0704172327 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 5 |
Hb_000958_090 |
0.0718431559 |
- |
- |
PREDICTED: cation/H(+) antiporter 28 [Jatropha curcas] |
| 6 |
Hb_000520_040 |
0.0767319146 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 7 |
Hb_001047_210 |
0.0785866777 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |
| 8 |
Hb_018591_020 |
0.0799476699 |
- |
- |
Ribulose bisphosphate carboxylase/oxygenase activase 1, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_000424_140 |
0.0812863805 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_002246_030 |
0.0815875892 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 11 |
Hb_000984_160 |
0.0828363784 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_003195_080 |
0.0841851573 |
- |
- |
PREDICTED: cytochrome P450 87A3-like [Jatropha curcas] |
| 13 |
Hb_001098_040 |
0.0854447742 |
- |
- |
hypothetical protein JCGZ_00749 [Jatropha curcas] |
| 14 |
Hb_005694_120 |
0.085740968 |
- |
- |
PREDICTED: calcium-transporting ATPase, endoplasmic reticulum-type [Jatropha curcas] |
| 15 |
Hb_173387_010 |
0.0868612256 |
- |
- |
hypothetical protein POPTR_0017s13190g, partial [Populus trichocarpa] |
| 16 |
Hb_025557_060 |
0.0873312163 |
transcription factor |
TF Family: MYB |
Myb-related protein 306, putative isoform 1 [Theobroma cacao] |
| 17 |
Hb_000046_120 |
0.0902357468 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 18 |
Hb_002182_010 |
0.0903275477 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase-like [Jatropha curcas] |
| 19 |
Hb_000500_240 |
0.0905909693 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000847_030 |
0.0906961619 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 2 [Jatropha curcas] |