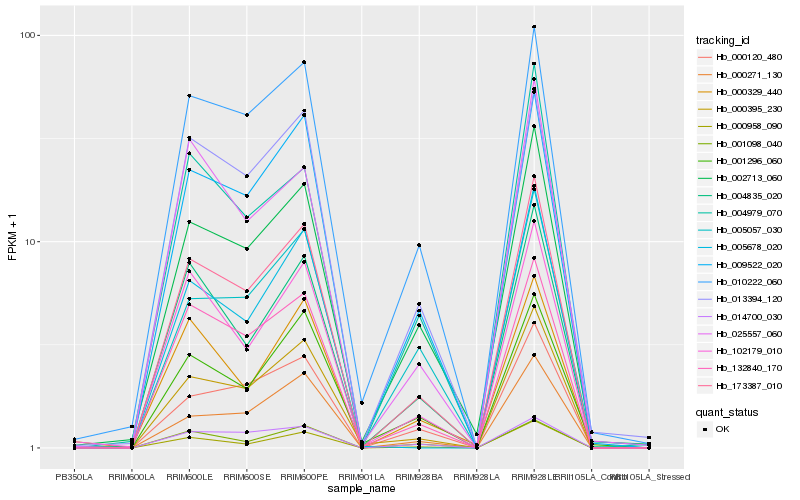
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_173387_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0017s13190g, partial [Populus trichocarpa] |
| 2 |
Hb_009522_020 |
0.0642482433 |
- |
- |
PREDICTED: protein ECERIFERUM 3 [Jatropha curcas] |
| 3 |
Hb_132840_170 |
0.0687155743 |
transcription factor |
TF Family: NAC |
Transcription factor [Theobroma cacao] |
| 4 |
Hb_000958_090 |
0.0763292993 |
- |
- |
PREDICTED: cation/H(+) antiporter 28 [Jatropha curcas] |
| 5 |
Hb_002713_060 |
0.0810374001 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.4 [Jatropha curcas] |
| 6 |
Hb_010222_060 |
0.0838823505 |
- |
- |
PREDICTED: acyltransferase-like protein At1g54570, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_004835_020 |
0.0868612256 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 8 |
Hb_000120_480 |
0.0890389811 |
- |
- |
- |
| 9 |
Hb_000395_230 |
0.0896307436 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Vitis vinifera] |
| 10 |
Hb_102179_010 |
0.0900670332 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_013394_120 |
0.0927816849 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 12 |
Hb_000271_130 |
0.1013341527 |
transcription factor |
TF Family: C2C2-Dof |
DNA binding with one finger 2.4, putative [Theobroma cacao] |
| 13 |
Hb_014700_030 |
0.1037512393 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 14 |
Hb_001296_060 |
0.1052585982 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 15 |
Hb_005678_020 |
0.1073259044 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_000329_440 |
0.1085260032 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 17 |
Hb_004979_070 |
0.1105297685 |
- |
- |
wall-associated receptor kinase galacturonan-binding protein [Medicago truncatula] |
| 18 |
Hb_001098_040 |
0.111355237 |
- |
- |
hypothetical protein JCGZ_00749 [Jatropha curcas] |
| 19 |
Hb_025557_060 |
0.1116982469 |
transcription factor |
TF Family: MYB |
Myb-related protein 306, putative isoform 1 [Theobroma cacao] |
| 20 |
Hb_005057_030 |
0.112579277 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |