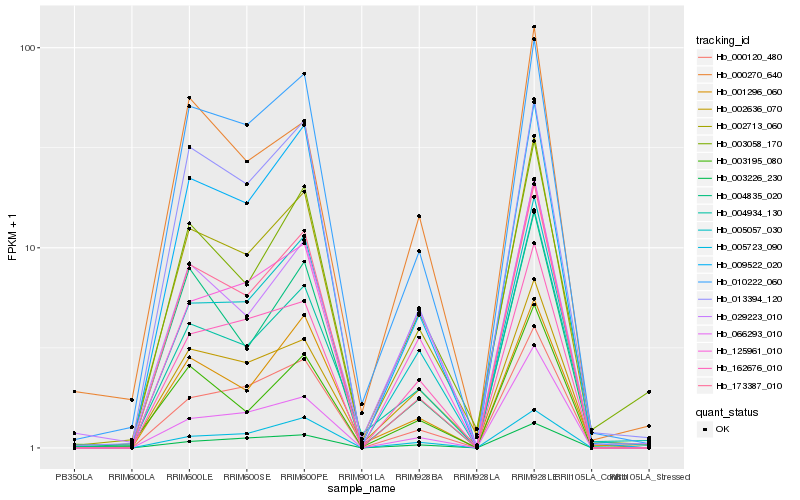
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002713_060 |
0.0 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.4 [Jatropha curcas] |
| 2 |
Hb_005057_030 |
0.0790134897 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 3 |
Hb_173387_010 |
0.0810374001 |
- |
- |
hypothetical protein POPTR_0017s13190g, partial [Populus trichocarpa] |
| 4 |
Hb_010222_060 |
0.0836692678 |
- |
- |
PREDICTED: acyltransferase-like protein At1g54570, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_002636_070 |
0.088578655 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 6 |
Hb_000120_480 |
0.090502065 |
- |
- |
- |
| 7 |
Hb_009522_020 |
0.0910132188 |
- |
- |
PREDICTED: protein ECERIFERUM 3 [Jatropha curcas] |
| 8 |
Hb_003058_170 |
0.0940873611 |
- |
- |
PREDICTED: F-box protein At2g26850 isoform X1 [Jatropha curcas] |
| 9 |
Hb_162676_010 |
0.0971907326 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_125961_010 |
0.0976696267 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_004934_130 |
0.0977099597 |
- |
- |
12-oxophytodienoate reductase 2 isoform 1 [Theobroma cacao] |
| 12 |
Hb_003195_080 |
0.100334717 |
- |
- |
PREDICTED: cytochrome P450 87A3-like [Jatropha curcas] |
| 13 |
Hb_029223_010 |
0.1019444545 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |
| 14 |
Hb_001296_060 |
0.1042719291 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 15 |
Hb_004835_020 |
0.1049528449 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 16 |
Hb_003226_230 |
0.1049700272 |
- |
- |
PREDICTED: transposon Ty3-G Gag-Pol polyprotein [Phoenix dactylifera] |
| 17 |
Hb_013394_120 |
0.1086054578 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 18 |
Hb_066293_010 |
0.1086738424 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 19 |
Hb_000270_640 |
0.1088689156 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 20 |
Hb_005723_090 |
0.1108757491 |
- |
- |
conserved hypothetical protein [Ricinus communis] |