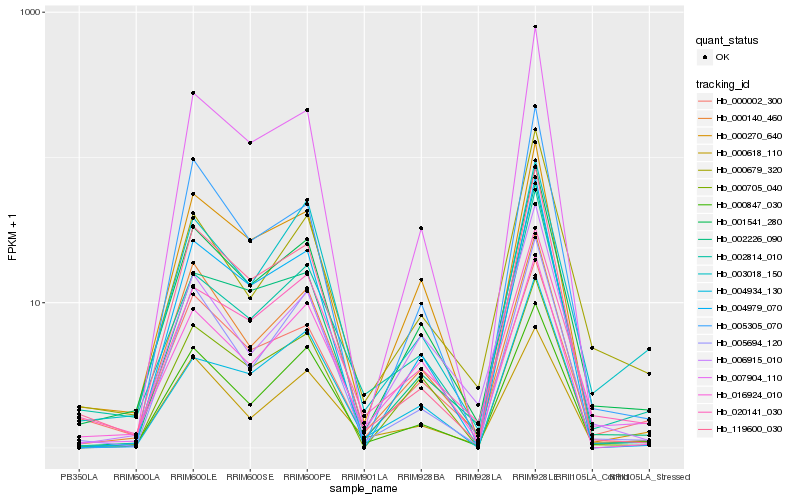
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001541_280 |
0.0 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 2 |
Hb_000270_640 |
0.0821011605 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 3 |
Hb_000847_030 |
0.0844630769 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 2 [Jatropha curcas] |
| 4 |
Hb_002814_010 |
0.0935809608 |
- |
- |
PREDICTED: adenylate kinase 5, chloroplastic [Jatropha curcas] |
| 5 |
Hb_020141_030 |
0.1004823039 |
- |
- |
solanesyl diphosphate synthase, putative [Ricinus communis] |
| 6 |
Hb_000140_460 |
0.101387918 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 7 |
Hb_000002_300 |
0.1024431526 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 8 |
Hb_006915_010 |
0.104354516 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 9 |
Hb_119600_030 |
0.1050434067 |
- |
- |
PREDICTED: uncharacterized protein LOC105635187 [Jatropha curcas] |
| 10 |
Hb_004979_070 |
0.1058152819 |
- |
- |
wall-associated receptor kinase galacturonan-binding protein [Medicago truncatula] |
| 11 |
Hb_005694_120 |
0.1067784541 |
- |
- |
PREDICTED: calcium-transporting ATPase, endoplasmic reticulum-type [Jatropha curcas] |
| 12 |
Hb_000679_320 |
0.1072076311 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105136146 [Populus euphratica] |
| 13 |
Hb_007904_110 |
0.107630732 |
- |
- |
PREDICTED: photosystem II core complex proteins psbY, chloroplastic-like [Populus euphratica] |
| 14 |
Hb_000705_040 |
0.1086634301 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000618_110 |
0.111619446 |
- |
- |
hypothetical protein POPTR_0003s09870g [Populus trichocarpa] |
| 16 |
Hb_002226_090 |
0.1118817256 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 17 |
Hb_016924_010 |
0.1125387082 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003018_150 |
0.1134184613 |
- |
- |
PREDICTED: uncharacterized protein LOC105628869 [Jatropha curcas] |
| 19 |
Hb_004934_130 |
0.1145815694 |
- |
- |
12-oxophytodienoate reductase 2 isoform 1 [Theobroma cacao] |
| 20 |
Hb_005305_070 |
0.1157574034 |
- |
- |
hypothetical protein POPTR_0001s26540g [Populus trichocarpa] |