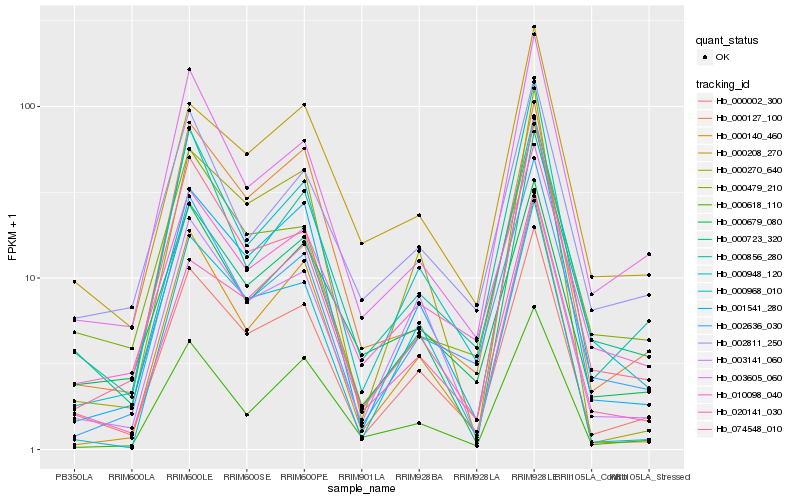
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000002_300 |
0.0 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 2 |
Hb_003141_060 |
0.0987981612 |
transcription factor |
TF Family: TRAF |
PREDICTED: ETO1-like protein 1 [Jatropha curcas] |
| 3 |
Hb_001541_280 |
0.1024431526 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 4 |
Hb_074548_010 |
0.1077196214 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_003605_060 |
0.1083258626 |
- |
- |
PREDICTED: ATP synthase subunit delta, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000948_120 |
0.1108707281 |
- |
- |
PREDICTED: uncharacterized protein LOC105634688 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002636_030 |
0.1111075423 |
- |
- |
hypothetical protein JCGZ_11800 [Jatropha curcas] |
| 8 |
Hb_010098_040 |
0.1113443602 |
- |
- |
PREDICTED: DNA-binding protein SMUBP-2 [Jatropha curcas] |
| 9 |
Hb_000968_010 |
0.1120031576 |
- |
- |
PREDICTED: glutamate receptor 3.7 isoform X1 [Jatropha curcas] |
| 10 |
Hb_020141_030 |
0.1129106542 |
- |
- |
solanesyl diphosphate synthase, putative [Ricinus communis] |
| 11 |
Hb_000618_110 |
0.1151523675 |
- |
- |
hypothetical protein POPTR_0003s09870g [Populus trichocarpa] |
| 12 |
Hb_000856_280 |
0.1153842763 |
- |
- |
PREDICTED: uncharacterized protein LOC105640491 [Jatropha curcas] |
| 13 |
Hb_000270_640 |
0.1157915518 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 14 |
Hb_000140_460 |
0.1169735152 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 15 |
Hb_000208_270 |
0.1196357516 |
- |
- |
PREDICTED: malate dehydrogenase [NADP], chloroplastic [Jatropha curcas] |
| 16 |
Hb_000679_080 |
0.1212472315 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000479_210 |
0.1225612491 |
- |
- |
PREDICTED: uncharacterized protein LOC105644126 [Jatropha curcas] |
| 18 |
Hb_000127_100 |
0.1228105623 |
- |
- |
PREDICTED: myb-related transcription factor, partner of profilin-like [Populus euphratica] |
| 19 |
Hb_000723_320 |
0.1270843866 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 [Jatropha curcas] |
| 20 |
Hb_002811_250 |
0.1274673674 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |