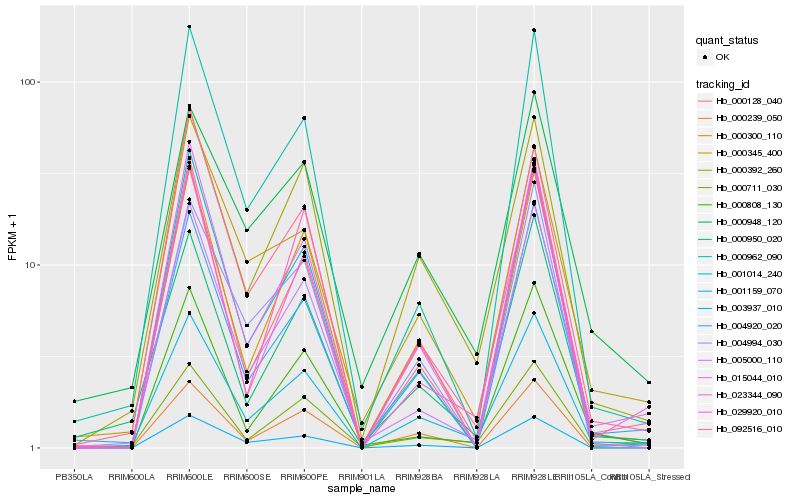
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001159_070 |
0.0 |
- |
- |
DNA photolyase, putative [Ricinus communis] |
| 2 |
Hb_000392_260 |
0.0725609087 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1B [Jatropha curcas] |
| 3 |
Hb_000711_030 |
0.093767 |
- |
- |
hypothetical protein POPTR_0019s02860g [Populus trichocarpa] |
| 4 |
Hb_004920_020 |
0.0984667259 |
- |
- |
hypothetical protein POPTR_0014s11700g [Populus trichocarpa] |
| 5 |
Hb_000950_020 |
0.0995523044 |
- |
- |
PREDICTED: glycerol-3-phosphate dehydrogenase [NAD(+)] GPDHC1, cytosolic [Jatropha curcas] |
| 6 |
Hb_005000_110 |
0.1004788546 |
- |
- |
PREDICTED: uncharacterized protein LOC105637872 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000948_120 |
0.104055508 |
- |
- |
PREDICTED: uncharacterized protein LOC105634688 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000300_110 |
0.1061873246 |
- |
- |
hypothetical protein JCGZ_02651 [Jatropha curcas] |
| 9 |
Hb_004994_030 |
0.1161645745 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |
| 10 |
Hb_023344_090 |
0.1166674739 |
- |
- |
Photosystem II reaction center W protein, putative [Ricinus communis] |
| 11 |
Hb_092516_010 |
0.119117585 |
- |
- |
hypothetical protein JCGZ_12248 [Jatropha curcas] |
| 12 |
Hb_000239_050 |
0.1210803642 |
- |
- |
PREDICTED: cyclin-D5-1-like [Jatropha curcas] |
| 13 |
Hb_000962_090 |
0.1216828105 |
- |
- |
omega-6 fatty acid desaturase [Hevea brasiliensis] |
| 14 |
Hb_000345_400 |
0.1219935576 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003937_010 |
0.1230291437 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |
| 16 |
Hb_015044_010 |
0.123313419 |
- |
- |
hypothetical protein JCGZ_25130 [Jatropha curcas] |
| 17 |
Hb_000128_040 |
0.1243087682 |
- |
- |
PREDICTED: uncharacterized protein LOC105641228 isoform X1 [Jatropha curcas] |
| 18 |
Hb_029920_010 |
0.1258317081 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 19 |
Hb_001014_240 |
0.1259187577 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.1 [Jatropha curcas] |
| 20 |
Hb_000808_130 |
0.1260214125 |
- |
- |
3-5 exonuclease, putative [Ricinus communis] |