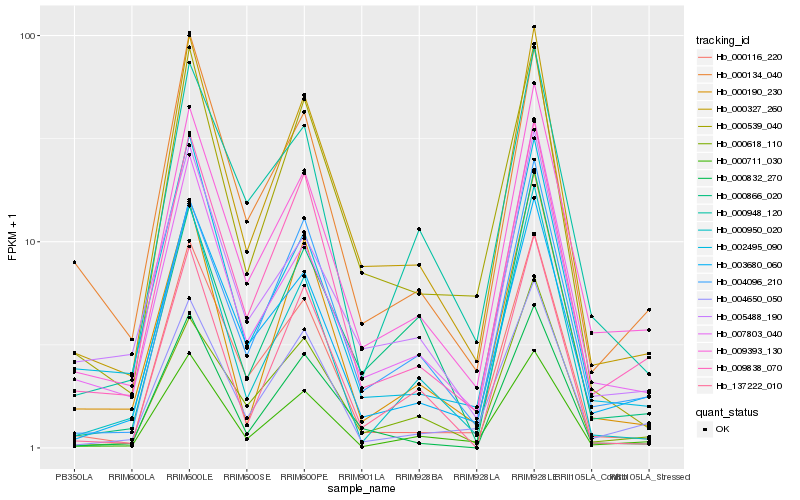
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000327_260 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000539_040 |
0.0708575214 |
- |
- |
PREDICTED: uncharacterized protein LOC105636110 isoform X1 [Jatropha curcas] |
| 3 |
Hb_009838_070 |
0.0807223377 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic [Jatropha curcas] |
| 4 |
Hb_009393_130 |
0.0922424297 |
- |
- |
PREDICTED: uncharacterized protein LOC105638053 isoform X4 [Jatropha curcas] |
| 5 |
Hb_000116_220 |
0.0930865943 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004096_210 |
0.0943433134 |
- |
- |
PREDICTED: uncharacterized protein LOC105638707 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000832_270 |
0.096816583 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000711_030 |
0.0988887705 |
- |
- |
hypothetical protein POPTR_0019s02860g [Populus trichocarpa] |
| 9 |
Hb_003680_060 |
0.1025026552 |
- |
- |
hypothetical protein VITISV_022322 [Vitis vinifera] |
| 10 |
Hb_137222_010 |
0.1065936634 |
- |
- |
PREDICTED: hippocampus abundant transcript-like protein 1 [Jatropha curcas] |
| 11 |
Hb_000618_110 |
0.1067406229 |
- |
- |
hypothetical protein POPTR_0003s09870g [Populus trichocarpa] |
| 12 |
Hb_000134_040 |
0.1082336579 |
- |
- |
PREDICTED: 30S ribosomal protein 2, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000190_230 |
0.1134434524 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 14 |
Hb_002495_090 |
0.1143395898 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 15 |
Hb_005488_190 |
0.1144665111 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 16 |
Hb_000866_020 |
0.1162306525 |
- |
- |
hypothetical protein B456_004G042000 [Gossypium raimondii] |
| 17 |
Hb_000950_020 |
0.1172156302 |
- |
- |
PREDICTED: glycerol-3-phosphate dehydrogenase [NAD(+)] GPDHC1, cytosolic [Jatropha curcas] |
| 18 |
Hb_007803_040 |
0.1175512546 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP37, chloroplastic [Jatropha curcas] |
| 19 |
Hb_004650_050 |
0.1178762065 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_000948_120 |
0.1241813629 |
- |
- |
PREDICTED: uncharacterized protein LOC105634688 isoform X1 [Jatropha curcas] |