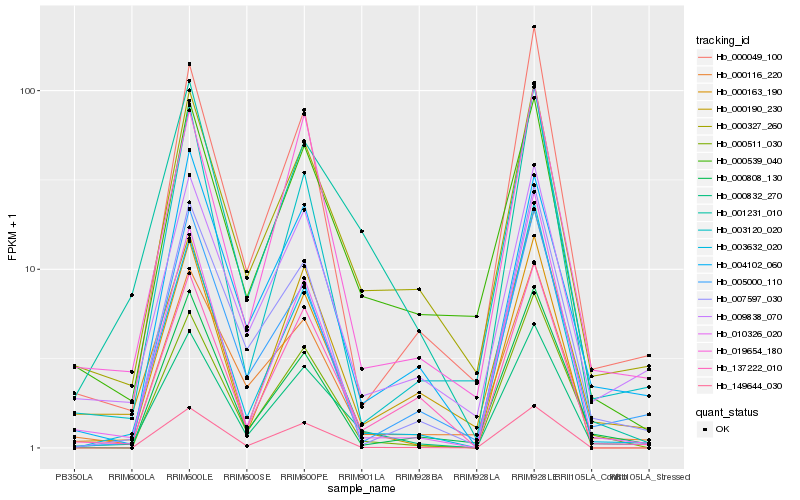
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000832_270 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000327_260 |
0.096816583 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001231_010 |
0.1089670442 |
- |
- |
calcium-binding family protein [Populus trichocarpa] |
| 4 |
Hb_000511_030 |
0.112793617 |
- |
- |
PREDICTED: uncharacterized protein LOC105636463 [Jatropha curcas] |
| 5 |
Hb_000163_190 |
0.1131230787 |
- |
- |
PREDICTED: 21 kDa protein [Populus euphratica] |
| 6 |
Hb_000116_220 |
0.1177502283 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_137222_010 |
0.1185971821 |
- |
- |
PREDICTED: hippocampus abundant transcript-like protein 1 [Jatropha curcas] |
| 8 |
Hb_009838_070 |
0.1192741846 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000539_040 |
0.1218597464 |
- |
- |
PREDICTED: uncharacterized protein LOC105636110 isoform X1 [Jatropha curcas] |
| 10 |
Hb_019654_180 |
0.1230704231 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 11 |
Hb_003632_020 |
0.1231183787 |
- |
- |
calcium-binding family protein [Populus trichocarpa] |
| 12 |
Hb_149644_030 |
0.1271671332 |
- |
- |
hypothetical protein POPTR_0005s18750g [Populus trichocarpa] |
| 13 |
Hb_000049_100 |
0.1297729494 |
- |
- |
Photosystem II reaction center W protein, putative [Ricinus communis] |
| 14 |
Hb_000808_130 |
0.1304711878 |
- |
- |
3-5 exonuclease, putative [Ricinus communis] |
| 15 |
Hb_007597_030 |
0.1316270498 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_003120_020 |
0.1318074711 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 13-like [Prunus mume] |
| 17 |
Hb_005000_110 |
0.1324144352 |
- |
- |
PREDICTED: uncharacterized protein LOC105637872 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000190_230 |
0.1337448345 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 19 |
Hb_004102_060 |
0.1340755953 |
- |
- |
JHL25H03.11 [Jatropha curcas] |
| 20 |
Hb_010326_020 |
0.1346308519 |
- |
- |
PREDICTED: thioredoxin-like 4, chloroplastic [Jatropha curcas] |