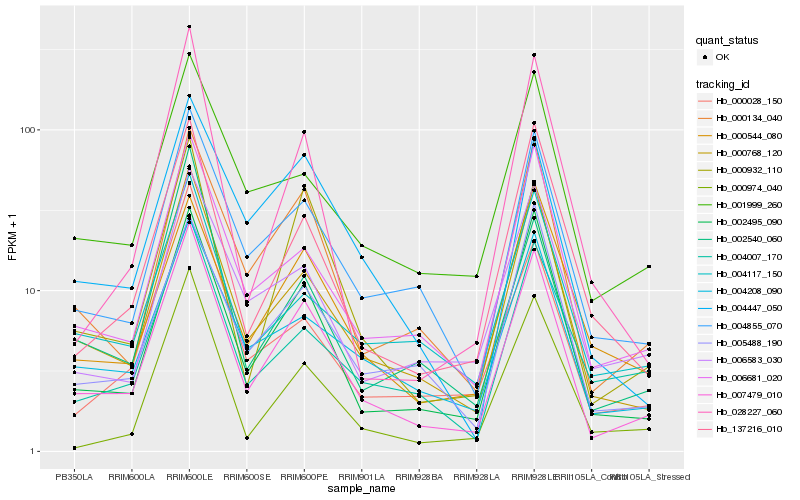
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007479_010 |
0.0 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 2 |
Hb_000768_120 |
0.0825048992 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigB [Jatropha curcas] |
| 3 |
Hb_002495_090 |
0.0902909292 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000134_040 |
0.0985614877 |
- |
- |
PREDICTED: 30S ribosomal protein 2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_004855_070 |
0.1088468246 |
- |
- |
PLASTID-SPECIFIC RIBOSOMAL protein 4 [Populus trichocarpa] |
| 6 |
Hb_006681_020 |
0.1112350837 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP38, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000932_110 |
0.1167517756 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 8 |
Hb_137216_010 |
0.1237448982 |
- |
- |
PREDICTED: unknown protein DS12 from 2D-PAGE of leaf, chloroplastic-like isoform X2 [Citrus sinensis] |
| 9 |
Hb_004208_090 |
0.1295872338 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 10 |
Hb_000974_040 |
0.1323934764 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g49770 [Jatropha curcas] |
| 11 |
Hb_001999_260 |
0.1335179368 |
- |
- |
PREDICTED: 50S ribosomal protein L4, chloroplastic [Jatropha curcas] |
| 12 |
Hb_005488_190 |
0.1370759753 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 13 |
Hb_006583_030 |
0.1374228271 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like isoform X4 [Jatropha curcas] |
| 14 |
Hb_004007_170 |
0.1425555177 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 15 |
Hb_004117_150 |
0.14354775 |
- |
- |
PREDICTED: uncharacterized protein LOC105649105 [Jatropha curcas] |
| 16 |
Hb_004447_050 |
0.1441242203 |
- |
- |
PREDICTED: high-light-induced protein, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002540_060 |
0.1456884164 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000028_150 |
0.145938367 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 19 |
Hb_028227_060 |
0.1480052367 |
- |
- |
PREDICTED: non-specific lipid transfer protein GPI-anchored 1-like [Populus euphratica] |
| 20 |
Hb_000544_080 |
0.1480641516 |
- |
- |
PREDICTED: uncharacterized protein LOC105631062 isoform X3 [Jatropha curcas] |