| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
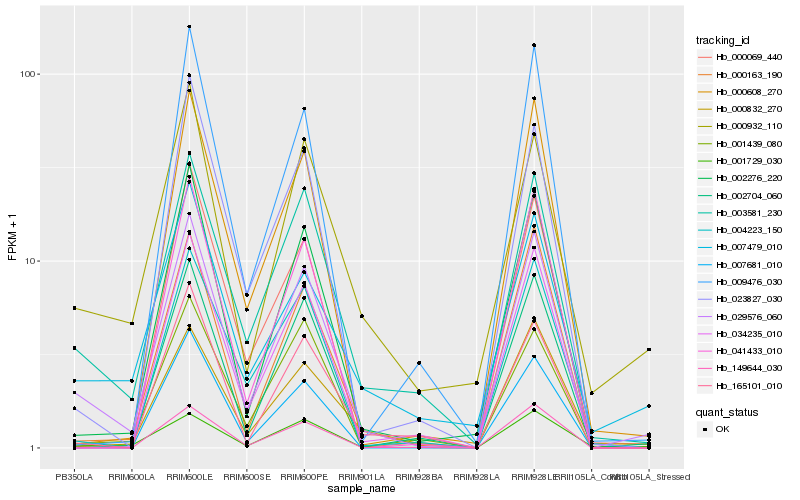
Hb_029576_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646366 [Jatropha curcas] |
| 2 |
Hb_003581_230 |
0.1031046789 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: SNF1-related protein kinase regulatory subunit gamma-1-like [Jatropha curcas] |
| 3 |
Hb_002276_220 |
0.1149759653 |
- |
- |
PREDICTED: uncharacterized protein LOC105640684 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000163_190 |
0.1247276077 |
- |
- |
PREDICTED: 21 kDa protein [Populus euphratica] |
| 5 |
Hb_034235_010 |
0.1362962594 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 6 |
Hb_000608_270 |
0.1365058509 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002704_060 |
0.1377631746 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 8 |
Hb_001729_030 |
0.138019617 |
- |
- |
hypothetical protein VITISV_044457 [Vitis vinifera] |
| 9 |
Hb_023827_030 |
0.1411538107 |
- |
- |
PREDICTED: thylakoid lumenal 16.5 kDa protein, chloroplastic-like [Populus euphratica] |
| 10 |
Hb_004223_150 |
0.1416436031 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Jatropha curcas] |
| 11 |
Hb_149644_030 |
0.1445199456 |
- |
- |
hypothetical protein POPTR_0005s18750g [Populus trichocarpa] |
| 12 |
Hb_000832_270 |
0.1448875851 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_041433_010 |
0.1449879369 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_000932_110 |
0.1455455029 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 15 |
Hb_165101_010 |
0.1457567004 |
- |
- |
PREDICTED: putative wall-associated receptor kinase-like 16 [Nelumbo nucifera] |
| 16 |
Hb_000069_440 |
0.1463867649 |
- |
- |
PREDICTED: uncharacterized protein LOC105642861 [Jatropha curcas] |
| 17 |
Hb_001439_080 |
0.1464533504 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 18 |
Hb_009476_030 |
0.1492181792 |
- |
- |
fatty acid desaturase [Manihot esculenta] |
| 19 |
Hb_007479_010 |
0.1495874427 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 20 |
Hb_007681_010 |
0.150947611 |
- |
- |
hypothetical protein RCOM_2128750 [Ricinus communis] |