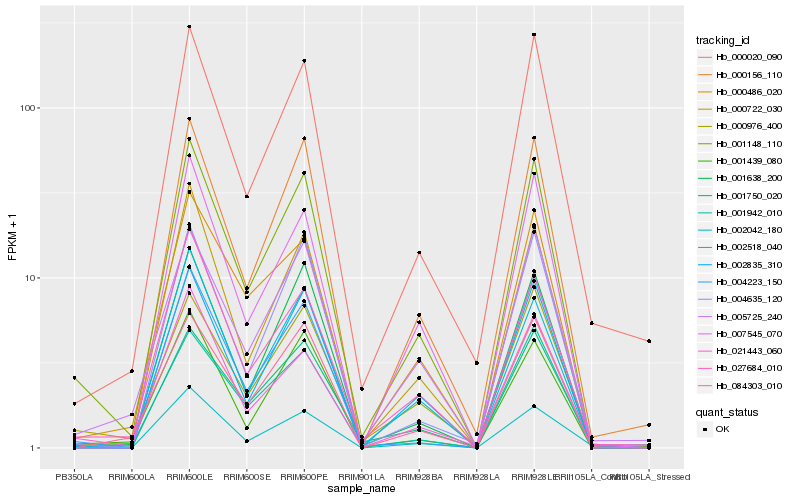
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001148_110 |
0.0 |
- |
- |
- |
| 2 |
Hb_000722_030 |
0.0904496716 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 3 |
Hb_001638_200 |
0.0935218848 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 43 [Jatropha curcas] |
| 4 |
Hb_002835_310 |
0.0938737831 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_000156_110 |
0.0941289796 |
- |
- |
Uncharacterized protein TCM_029490 [Theobroma cacao] |
| 6 |
Hb_001942_010 |
0.0955234584 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |
| 7 |
Hb_002042_180 |
0.0972806367 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 8 |
Hb_000486_020 |
0.0974233454 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.2 [Jatropha curcas] |
| 9 |
Hb_004223_150 |
0.0989328763 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Jatropha curcas] |
| 10 |
Hb_021443_060 |
0.0991910263 |
- |
- |
PTH-1 family protein [Populus trichocarpa] |
| 11 |
Hb_007545_070 |
0.1001443704 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 306 [Populus euphratica] |
| 12 |
Hb_000976_400 |
0.1007191049 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 13 |
Hb_000020_090 |
0.101538778 |
- |
- |
hypothetical protein JCGZ_09084 [Jatropha curcas] |
| 14 |
Hb_027684_010 |
0.1021472563 |
- |
- |
hypothetical protein RCOM_0504250 [Ricinus communis] |
| 15 |
Hb_002518_040 |
0.1034657408 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 16 |
Hb_001439_080 |
0.1045795494 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 17 |
Hb_005725_240 |
0.1052097668 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH74 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004635_120 |
0.1057151356 |
- |
- |
PREDICTED: rho GTPase-activating protein 4-like [Jatropha curcas] |
| 19 |
Hb_001750_020 |
0.1072568192 |
- |
- |
PREDICTED: MATE efflux family protein 6-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_084303_010 |
0.1076455905 |
- |
- |
conserved hypothetical protein [Ricinus communis] |