| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
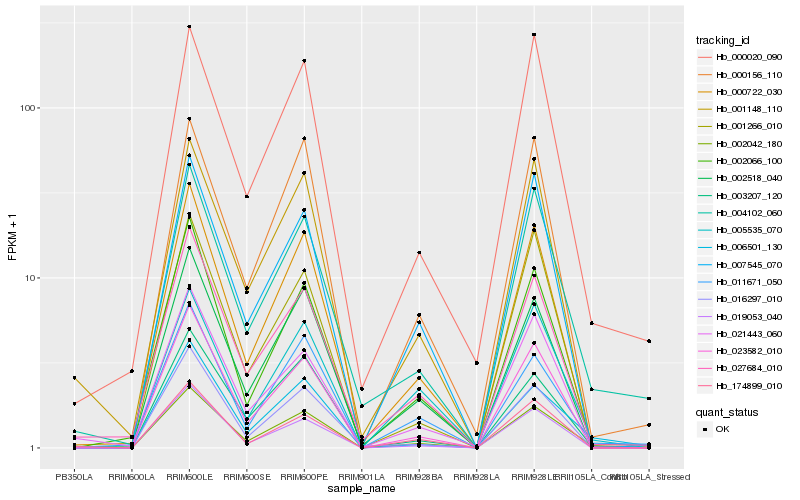
Hb_002042_180 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 2 |
Hb_000722_030 |
0.0921872596 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 3 |
Hb_001148_110 |
0.0972806367 |
- |
- |
- |
| 4 |
Hb_027684_010 |
0.0993013662 |
- |
- |
hypothetical protein RCOM_0504250 [Ricinus communis] |
| 5 |
Hb_023582_010 |
0.1011670356 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004102_060 |
0.101315283 |
- |
- |
JHL25H03.11 [Jatropha curcas] |
| 7 |
Hb_002518_040 |
0.1025264391 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 8 |
Hb_021443_060 |
0.1030996079 |
- |
- |
PTH-1 family protein [Populus trichocarpa] |
| 9 |
Hb_174899_010 |
0.1069552749 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 10 |
Hb_003207_120 |
0.1088440397 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 11 |
Hb_002066_100 |
0.1123937138 |
- |
- |
PREDICTED: protein disulfide-isomerase SCO2 [Jatropha curcas] |
| 12 |
Hb_000020_090 |
0.1130151377 |
- |
- |
hypothetical protein JCGZ_09084 [Jatropha curcas] |
| 13 |
Hb_006501_130 |
0.1137640057 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_016297_010 |
0.113883863 |
- |
- |
PREDICTED: uncharacterized protein LOC105638665 [Jatropha curcas] |
| 15 |
Hb_000156_110 |
0.1140789561 |
- |
- |
Uncharacterized protein TCM_029490 [Theobroma cacao] |
| 16 |
Hb_007545_070 |
0.1147804931 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 306 [Populus euphratica] |
| 17 |
Hb_005535_070 |
0.1158051162 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_019053_040 |
0.1164557408 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_011671_050 |
0.1171181603 |
- |
- |
PREDICTED: F-box protein At5g07670 [Jatropha curcas] |
| 20 |
Hb_001266_010 |
0.1180342679 |
- |
- |
PREDICTED: uncharacterized protein LOC101206474 [Cucumis sativus] |