| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
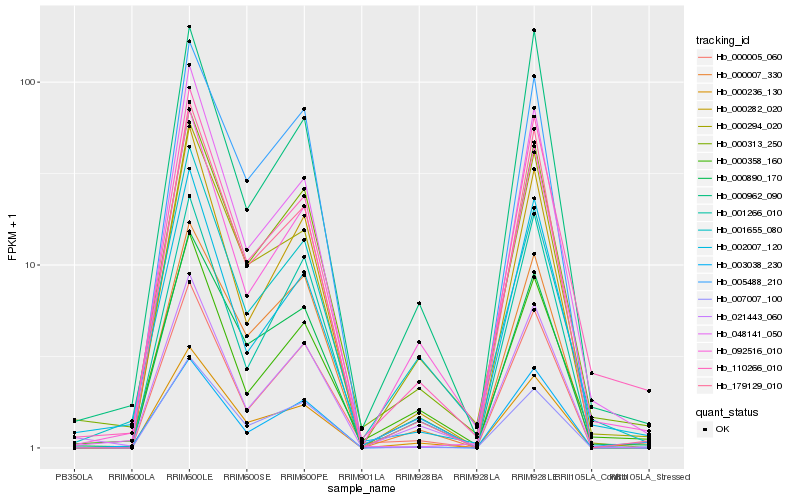
Hb_000313_250 |
0.0 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 2 |
Hb_000890_170 |
0.0747309918 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 5 [Jatropha curcas] |
| 3 |
Hb_110266_010 |
0.0778792913 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g21222-like [Jatropha curcas] |
| 4 |
Hb_092516_010 |
0.0789182096 |
- |
- |
hypothetical protein JCGZ_12248 [Jatropha curcas] |
| 5 |
Hb_002007_120 |
0.0833882689 |
- |
- |
PREDICTED: psbP domain-containing protein 7, chloroplastic [Jatropha curcas] |
| 6 |
Hb_021443_060 |
0.0836552346 |
- |
- |
PTH-1 family protein [Populus trichocarpa] |
| 7 |
Hb_000005_060 |
0.087558064 |
- |
- |
PREDICTED: uncharacterized protein At1g08160 [Jatropha curcas] |
| 8 |
Hb_000358_160 |
0.0888294899 |
- |
- |
PREDICTED: uncharacterized protein LOC105637769 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000294_020 |
0.0915951965 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 10 |
Hb_007007_100 |
0.0920007483 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.1 [Jatropha curcas] |
| 11 |
Hb_000236_130 |
0.0942512334 |
- |
- |
PREDICTED: protein ECERIFERUM 26 [Jatropha curcas] |
| 12 |
Hb_005488_210 |
0.0947043821 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 13 |
Hb_000007_330 |
0.0957073723 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 14 |
Hb_000282_020 |
0.0966616935 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003038_230 |
0.0981777547 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 7 [Jatropha curcas] |
| 16 |
Hb_179129_010 |
0.0990656598 |
- |
- |
hypothetical protein POPTR_0018s10470g [Populus trichocarpa] |
| 17 |
Hb_000962_090 |
0.100515687 |
- |
- |
omega-6 fatty acid desaturase [Hevea brasiliensis] |
| 18 |
Hb_001266_010 |
0.1013052653 |
- |
- |
PREDICTED: uncharacterized protein LOC101206474 [Cucumis sativus] |
| 19 |
Hb_001655_080 |
0.102200936 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 20 |
Hb_048141_050 |
0.1022870145 |
- |
- |
hypothetical protein PRUPE_ppa009153mg [Prunus persica] |