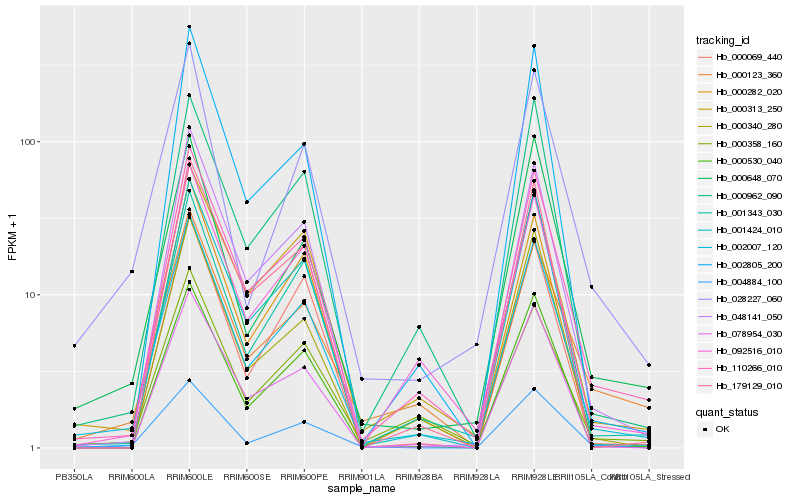
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002007_120 |
0.0 |
- |
- |
PREDICTED: psbP domain-containing protein 7, chloroplastic [Jatropha curcas] |
| 2 |
Hb_110266_010 |
0.0613355731 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g21222-like [Jatropha curcas] |
| 3 |
Hb_000313_250 |
0.0833882689 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 4 |
Hb_048141_050 |
0.0840674374 |
- |
- |
hypothetical protein PRUPE_ppa009153mg [Prunus persica] |
| 5 |
Hb_092516_010 |
0.0905678996 |
- |
- |
hypothetical protein JCGZ_12248 [Jatropha curcas] |
| 6 |
Hb_000530_040 |
0.0929663499 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000962_090 |
0.0931936898 |
- |
- |
omega-6 fatty acid desaturase [Hevea brasiliensis] |
| 8 |
Hb_000358_160 |
0.0945948235 |
- |
- |
PREDICTED: uncharacterized protein LOC105637769 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001424_010 |
0.0973336337 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 10 |
Hb_078954_030 |
0.0976229485 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001343_030 |
0.0993763353 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit N, chloroplastic [Jatropha curcas] |
| 12 |
Hb_179129_010 |
0.1001667275 |
- |
- |
hypothetical protein POPTR_0018s10470g [Populus trichocarpa] |
| 13 |
Hb_004884_100 |
0.1002648671 |
- |
- |
PREDICTED: lectin-domain containing receptor kinase VI.4-like [Jatropha curcas] |
| 14 |
Hb_028227_060 |
0.1025671444 |
- |
- |
PREDICTED: non-specific lipid transfer protein GPI-anchored 1-like [Populus euphratica] |
| 15 |
Hb_000340_280 |
0.1025722989 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002805_200 |
0.1037837676 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 17 |
Hb_000282_020 |
0.1041344002 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000069_440 |
0.1049620625 |
- |
- |
PREDICTED: uncharacterized protein LOC105642861 [Jatropha curcas] |
| 19 |
Hb_000648_070 |
0.1050696374 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 20 |
Hb_000123_360 |
0.105469734 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 1, chloroplastic isoform X1 [Jatropha curcas] |