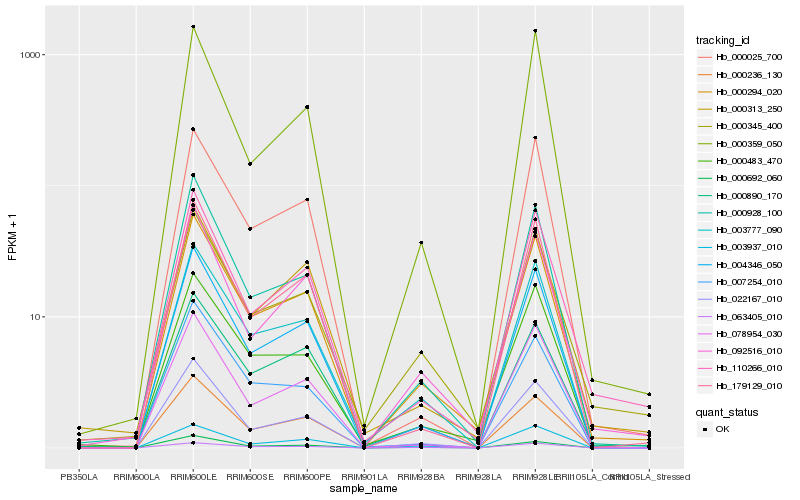
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000294_020 |
0.0 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 2 |
Hb_003777_090 |
0.0571689171 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 3 |
Hb_000483_470 |
0.067766999 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000236_130 |
0.0693610594 |
- |
- |
PREDICTED: protein ECERIFERUM 26 [Jatropha curcas] |
| 5 |
Hb_092516_010 |
0.0710697373 |
- |
- |
hypothetical protein JCGZ_12248 [Jatropha curcas] |
| 6 |
Hb_000928_100 |
0.0741101233 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 7 |
Hb_022167_010 |
0.0764559941 |
- |
- |
Kinase, putative isoform 2 [Theobroma cacao] |
| 8 |
Hb_000345_400 |
0.0816514379 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000890_170 |
0.0841687912 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 5 [Jatropha curcas] |
| 10 |
Hb_000692_060 |
0.0847892555 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 11 |
Hb_110266_010 |
0.085055299 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g21222-like [Jatropha curcas] |
| 12 |
Hb_179129_010 |
0.0882412257 |
- |
- |
hypothetical protein POPTR_0018s10470g [Populus trichocarpa] |
| 13 |
Hb_000313_250 |
0.0915951965 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 14 |
Hb_003937_010 |
0.0918455381 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |
| 15 |
Hb_078954_030 |
0.092780451 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_007254_010 |
0.0946129434 |
- |
- |
hypothetical protein JCGZ_25214 [Jatropha curcas] |
| 17 |
Hb_004346_050 |
0.0952098092 |
- |
- |
PREDICTED: ABC transporter G family member 22 [Jatropha curcas] |
| 18 |
Hb_063405_010 |
0.0965154644 |
- |
- |
PREDICTED: probable terpene synthase 3 [Jatropha curcas] |
| 19 |
Hb_000025_700 |
0.0988992666 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 20 |
Hb_000359_050 |
0.1002715678 |
- |
- |
Oxygen-evolving enhancer protein 2, chloroplast precursor, putative [Ricinus communis] |