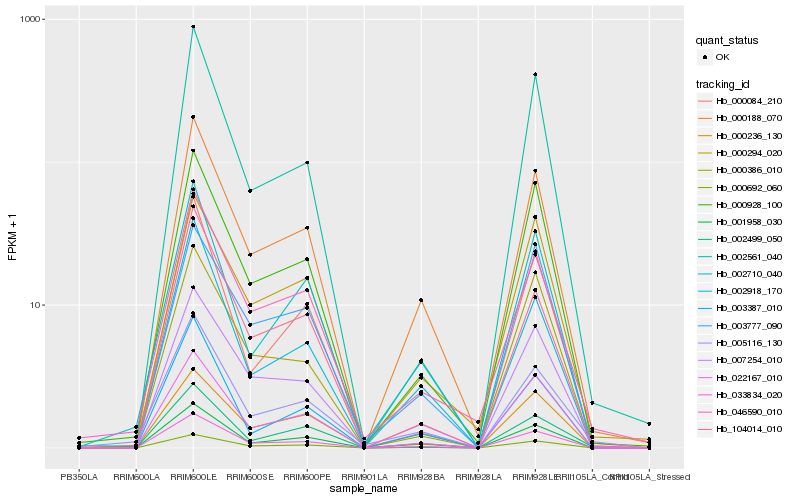
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033834_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000188_070 |
0.0466382073 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 3 |
Hb_000692_060 |
0.0512170677 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 4 |
Hb_007254_010 |
0.054906783 |
- |
- |
hypothetical protein JCGZ_25214 [Jatropha curcas] |
| 5 |
Hb_022167_010 |
0.0720695365 |
- |
- |
Kinase, putative isoform 2 [Theobroma cacao] |
| 6 |
Hb_000928_100 |
0.0745296519 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 7 |
Hb_001958_030 |
0.075664357 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 8 |
Hb_002561_040 |
0.0925942982 |
- |
- |
Photosynthetic electron transfer C [Theobroma cacao] |
| 9 |
Hb_000236_130 |
0.094215167 |
- |
- |
PREDICTED: protein ECERIFERUM 26 [Jatropha curcas] |
| 10 |
Hb_000386_010 |
0.0987454849 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002710_040 |
0.1008268482 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |
| 12 |
Hb_046590_010 |
0.1042609078 |
- |
- |
Cycloeucalenol cycloisomerase, putative [Ricinus communis] |
| 13 |
Hb_002918_170 |
0.1046331005 |
- |
- |
PREDICTED: MACPF domain-containing protein NSL1 [Jatropha curcas] |
| 14 |
Hb_104014_010 |
0.1051415291 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39020 isoform X2 [Jatropha curcas] |
| 15 |
Hb_003387_010 |
0.1072661859 |
- |
- |
PREDICTED: protein kinase PINOID 2 [Gossypium raimondii] |
| 16 |
Hb_000084_210 |
0.1084954438 |
- |
- |
hypothetical protein POPTR_0016s08050g [Populus trichocarpa] |
| 17 |
Hb_000294_020 |
0.1113247742 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 18 |
Hb_003777_090 |
0.1134905536 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 19 |
Hb_005116_130 |
0.1152398155 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa a, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002499_050 |
0.1154281949 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |