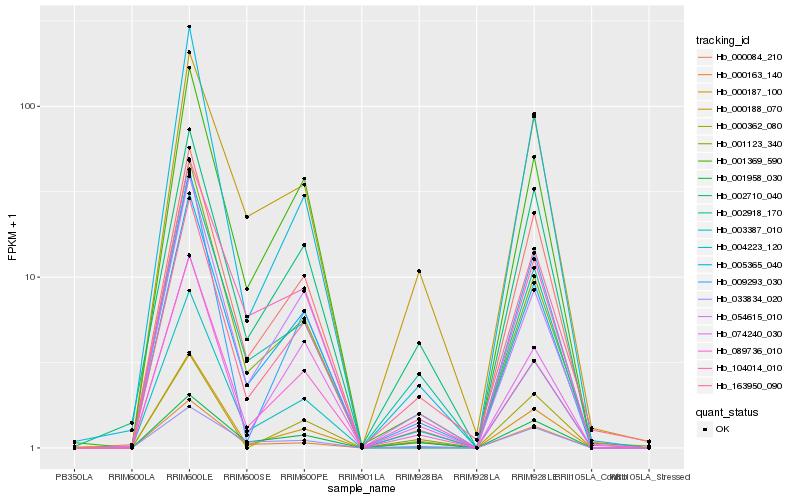
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003387_010 |
0.0 |
- |
- |
PREDICTED: protein kinase PINOID 2 [Gossypium raimondii] |
| 2 |
Hb_002918_170 |
0.0508764514 |
- |
- |
PREDICTED: MACPF domain-containing protein NSL1 [Jatropha curcas] |
| 3 |
Hb_002710_040 |
0.0880158272 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |
| 4 |
Hb_089736_010 |
0.0948403728 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 7 [Populus euphratica] |
| 5 |
Hb_001958_030 |
0.0950569147 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 6 |
Hb_004223_120 |
0.0966576067 |
- |
- |
PREDICTED: protein SPA, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000084_210 |
0.0997614226 |
- |
- |
hypothetical protein POPTR_0016s08050g [Populus trichocarpa] |
| 8 |
Hb_000188_070 |
0.1041938498 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 9 |
Hb_054615_010 |
0.1064466619 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |
| 10 |
Hb_033834_020 |
0.1072661859 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_009293_030 |
0.1102941179 |
- |
- |
PREDICTED: salicylate carboxymethyltransferase [Jatropha curcas] |
| 12 |
Hb_163950_090 |
0.1116067429 |
- |
- |
PREDICTED: traB domain-containing protein isoform X1 [Jatropha curcas] |
| 13 |
Hb_000163_140 |
0.1117096103 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 14 |
Hb_001369_590 |
0.113738402 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s14600g [Populus trichocarpa] |
| 15 |
Hb_000187_100 |
0.1174353912 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 16 |
Hb_001123_340 |
0.1184007428 |
- |
- |
PREDICTED: uncharacterized protein LOC105123089 isoform X1 [Populus euphratica] |
| 17 |
Hb_005365_040 |
0.1189244237 |
- |
- |
hypothetical protein JCGZ_15100 [Jatropha curcas] |
| 18 |
Hb_104014_010 |
0.120015723 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39020 isoform X2 [Jatropha curcas] |
| 19 |
Hb_074240_030 |
0.122217261 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 3-like [Jatropha curcas] |
| 20 |
Hb_000362_080 |
0.1267152955 |
- |
- |
hypothetical protein EUTSA_v10027329mg, partial [Eutrema salsugineum] |