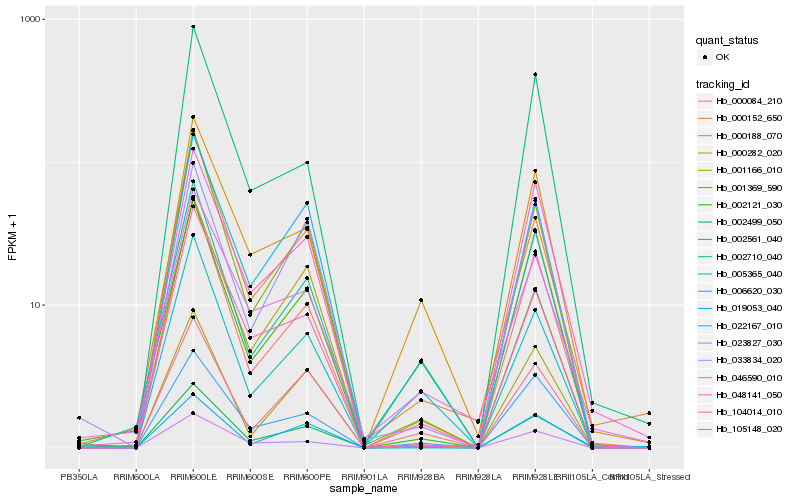
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002499_050 |
0.0 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 2 |
Hb_000084_210 |
0.0734149795 |
- |
- |
hypothetical protein POPTR_0016s08050g [Populus trichocarpa] |
| 3 |
Hb_001369_590 |
0.0751668284 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s14600g [Populus trichocarpa] |
| 4 |
Hb_006620_030 |
0.0806618975 |
- |
- |
nitrate reductase, putative [Ricinus communis] |
| 5 |
Hb_000282_020 |
0.0903221951 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_005365_040 |
0.0942062528 |
- |
- |
hypothetical protein JCGZ_15100 [Jatropha curcas] |
| 7 |
Hb_048141_050 |
0.0943266401 |
- |
- |
hypothetical protein PRUPE_ppa009153mg [Prunus persica] |
| 8 |
Hb_105148_020 |
0.0949194575 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase [Jatropha curcas] |
| 9 |
Hb_000152_650 |
0.1000121805 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 10 |
Hb_001166_010 |
0.106594245 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 11 |
Hb_104014_010 |
0.106952017 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39020 isoform X2 [Jatropha curcas] |
| 12 |
Hb_019053_040 |
0.1079801675 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 13 |
Hb_022167_010 |
0.1133905906 |
- |
- |
Kinase, putative isoform 2 [Theobroma cacao] |
| 14 |
Hb_002561_040 |
0.113616304 |
- |
- |
Photosynthetic electron transfer C [Theobroma cacao] |
| 15 |
Hb_002121_030 |
0.1150085784 |
- |
- |
Ribonuclease P protein subunit P38-related isoform 1 [Theobroma cacao] |
| 16 |
Hb_002710_040 |
0.1152918798 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |
| 17 |
Hb_033834_020 |
0.1154281949 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_023827_030 |
0.1169846345 |
- |
- |
PREDICTED: thylakoid lumenal 16.5 kDa protein, chloroplastic-like [Populus euphratica] |
| 19 |
Hb_046590_010 |
0.1171067557 |
- |
- |
Cycloeucalenol cycloisomerase, putative [Ricinus communis] |
| 20 |
Hb_000188_070 |
0.1180520948 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |