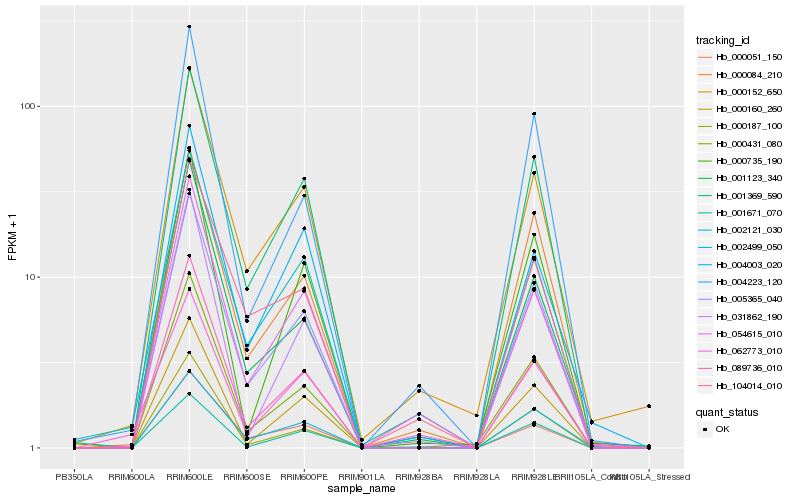
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005365_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_15100 [Jatropha curcas] |
| 2 |
Hb_001369_590 |
0.0477412907 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s14600g [Populus trichocarpa] |
| 3 |
Hb_000051_150 |
0.0735932426 |
- |
- |
hypothetical protein CICLE_v10003316mg [Citrus clementina] |
| 4 |
Hb_000187_100 |
0.0779941829 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 5 |
Hb_000160_260 |
0.0784133003 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 6 |
Hb_002121_030 |
0.0811755253 |
- |
- |
Ribonuclease P protein subunit P38-related isoform 1 [Theobroma cacao] |
| 7 |
Hb_000084_210 |
0.0843744618 |
- |
- |
hypothetical protein POPTR_0016s08050g [Populus trichocarpa] |
| 8 |
Hb_000431_080 |
0.0862855127 |
- |
- |
PREDICTED: uncharacterized protein LOC105632547 [Jatropha curcas] |
| 9 |
Hb_000152_650 |
0.0907906776 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 10 |
Hb_001123_340 |
0.0923794173 |
- |
- |
PREDICTED: uncharacterized protein LOC105123089 isoform X1 [Populus euphratica] |
| 11 |
Hb_002499_050 |
0.0942062528 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 12 |
Hb_054615_010 |
0.0969801673 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |
| 13 |
Hb_001671_070 |
0.0984298444 |
- |
- |
kinase, putative [Ricinus communis] |
| 14 |
Hb_104014_010 |
0.0992566746 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39020 isoform X2 [Jatropha curcas] |
| 15 |
Hb_089736_010 |
0.103091026 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 7 [Populus euphratica] |
| 16 |
Hb_004003_020 |
0.10331607 |
- |
- |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_000735_190 |
0.1044441289 |
- |
- |
hypothetical protein POPTR_0006s19340g [Populus trichocarpa] |
| 18 |
Hb_004223_120 |
0.1054219604 |
- |
- |
PREDICTED: protein SPA, chloroplastic [Jatropha curcas] |
| 19 |
Hb_062773_010 |
0.1054990837 |
- |
- |
beta glucosidase [Manihot esculenta] |
| 20 |
Hb_031862_190 |
0.1068671311 |
- |
- |
- |