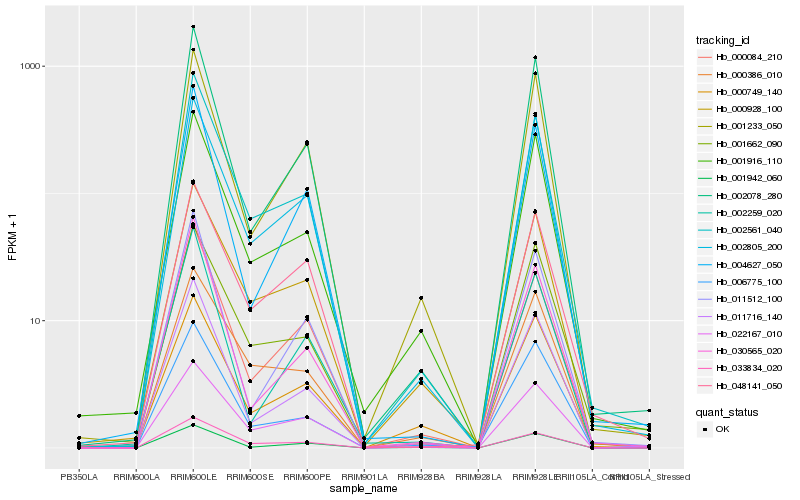
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002561_040 |
0.0 |
- |
- |
Photosynthetic electron transfer C [Theobroma cacao] |
| 2 |
Hb_000084_210 |
0.0712990684 |
- |
- |
hypothetical protein POPTR_0016s08050g [Populus trichocarpa] |
| 3 |
Hb_011716_140 |
0.0777896679 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |
| 4 |
Hb_000928_100 |
0.0849033517 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 5 |
Hb_002078_280 |
0.0871136637 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 6 |
Hb_000386_010 |
0.0877135841 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001916_110 |
0.0908202717 |
- |
- |
PREDICTED: uncharacterized protein LOC105641510 [Jatropha curcas] |
| 8 |
Hb_033834_020 |
0.0925942982 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_022167_010 |
0.0928903563 |
- |
- |
Kinase, putative isoform 2 [Theobroma cacao] |
| 10 |
Hb_030565_020 |
0.0951693638 |
- |
- |
PREDICTED: protein ECERIFERUM 26 [Jatropha curcas] |
| 11 |
Hb_006775_100 |
0.096870741 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_002805_200 |
0.0979617318 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 13 |
Hb_001662_090 |
0.1002581989 |
- |
- |
PREDICTED: thiosulfate sulfurtransferase 16, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_004627_050 |
0.1014482375 |
- |
- |
Chlorophyll a-b binding protein 6A [Morus notabilis] |
| 15 |
Hb_048141_050 |
0.1026323874 |
- |
- |
hypothetical protein PRUPE_ppa009153mg [Prunus persica] |
| 16 |
Hb_011512_100 |
0.1032902279 |
- |
- |
PREDICTED: uncharacterized protein LOC105650382 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001233_050 |
0.1050331864 |
- |
- |
Magnesium-protoporphyrin IX monomethyl ester [oxidative] cyclase, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_000749_140 |
0.1092381538 |
- |
- |
PREDICTED: 2-succinylbenzoate--CoA ligase, chloroplastic/peroxisomal [Jatropha curcas] |
| 19 |
Hb_001942_060 |
0.1098821913 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002259_020 |
0.1126836769 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |