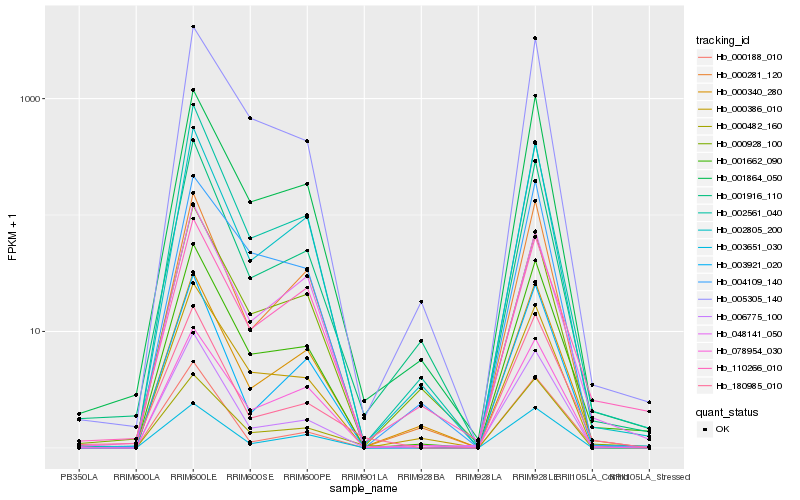
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001662_090 |
0.0 |
- |
- |
PREDICTED: thiosulfate sulfurtransferase 16, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_002805_200 |
0.0758311555 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 3 |
Hb_001864_050 |
0.0759547151 |
- |
- |
chloroplast oxygen-evolving enhancer protein [Manihot esculenta] |
| 4 |
Hb_005305_140 |
0.0810903004 |
- |
- |
unknown [Populus trichocarpa] |
| 5 |
Hb_006775_100 |
0.0846944529 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_001916_110 |
0.0860013841 |
- |
- |
PREDICTED: uncharacterized protein LOC105641510 [Jatropha curcas] |
| 7 |
Hb_000386_010 |
0.0871816529 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000340_280 |
0.0968328215 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002561_040 |
0.1002581989 |
- |
- |
Photosynthetic electron transfer C [Theobroma cacao] |
| 10 |
Hb_078954_030 |
0.1007856817 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000188_010 |
0.1032802546 |
- |
- |
PREDICTED: cytochrome P450 94C1-like [Jatropha curcas] |
| 12 |
Hb_000482_160 |
0.1037669669 |
- |
- |
hypothetical protein JCGZ_07670 [Jatropha curcas] |
| 13 |
Hb_000281_120 |
0.1040098832 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_000928_100 |
0.1040227729 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 15 |
Hb_003921_020 |
0.1041802813 |
- |
- |
hypothetical protein JCGZ_10628 [Jatropha curcas] |
| 16 |
Hb_048141_050 |
0.1071364742 |
- |
- |
hypothetical protein PRUPE_ppa009153mg [Prunus persica] |
| 17 |
Hb_110266_010 |
0.1084648474 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g21222-like [Jatropha curcas] |
| 18 |
Hb_003651_030 |
0.1088331218 |
- |
- |
lyase, putative [Ricinus communis] |
| 19 |
Hb_004109_140 |
0.1103476563 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_180985_010 |
0.1111222946 |
- |
- |
NDH-dependent cyclic electron flow 1 isoform 1 [Theobroma cacao] |