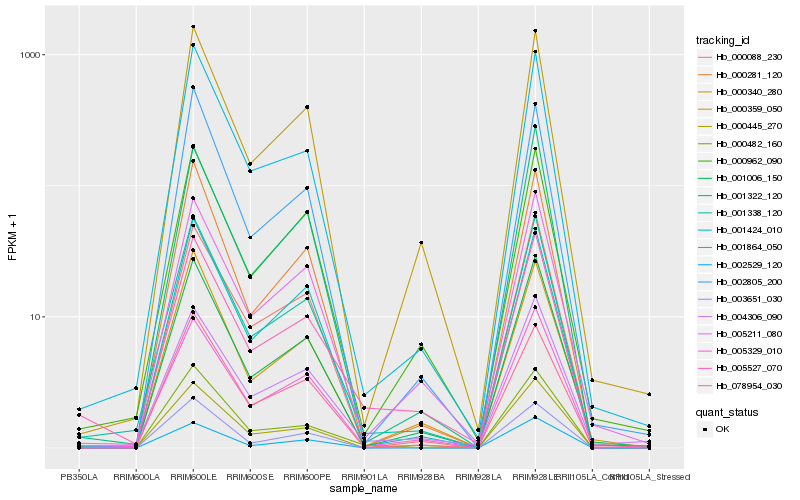
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001338_120 |
0.0 |
- |
- |
hypothetical chloroplast RF68 (chloroplast) [Ipomoea batatas] |
| 2 |
Hb_001864_050 |
0.0576282236 |
- |
- |
chloroplast oxygen-evolving enhancer protein [Manihot esculenta] |
| 3 |
Hb_001006_150 |
0.0611173471 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 4 |
Hb_005329_010 |
0.0663987388 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 [Jatropha curcas] |
| 5 |
Hb_000359_050 |
0.0705776092 |
- |
- |
Oxygen-evolving enhancer protein 2, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_000281_120 |
0.0733167303 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_000445_270 |
0.0735499409 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_003651_030 |
0.0742294746 |
- |
- |
lyase, putative [Ricinus communis] |
| 9 |
Hb_005211_080 |
0.0780277703 |
- |
- |
hypothetical protein POPTR_0010s13740g [Populus trichocarpa] |
| 10 |
Hb_078954_030 |
0.0782464239 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000962_090 |
0.0791158053 |
- |
- |
omega-6 fatty acid desaturase [Hevea brasiliensis] |
| 12 |
Hb_005527_070 |
0.0803839213 |
- |
- |
short-chain dehydrogenase/reductase family protein [Populus trichocarpa] |
| 13 |
Hb_000088_230 |
0.0812310926 |
- |
- |
hypothetical protein POPTR_0013s11340g [Populus trichocarpa] |
| 14 |
Hb_000482_160 |
0.0812785262 |
- |
- |
hypothetical protein JCGZ_07670 [Jatropha curcas] |
| 15 |
Hb_001424_010 |
0.0819160879 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 16 |
Hb_004306_090 |
0.0826656966 |
- |
- |
PREDICTED: uncharacterized protein LOC105644392 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000340_280 |
0.0840225678 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002805_200 |
0.0847860625 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 19 |
Hb_001322_120 |
0.0892004045 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 20 |
Hb_002529_120 |
0.0911160193 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 7-like [Jatropha curcas] |