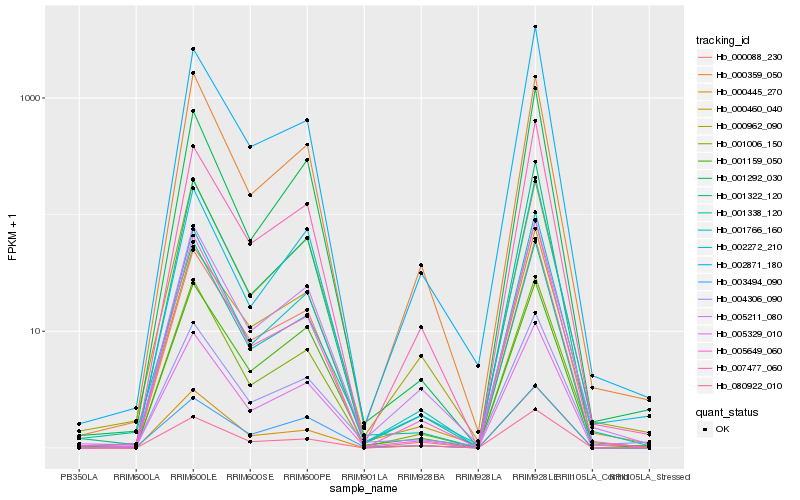
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005329_010 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 [Jatropha curcas] |
| 2 |
Hb_005211_080 |
0.0511226133 |
- |
- |
hypothetical protein POPTR_0010s13740g [Populus trichocarpa] |
| 3 |
Hb_001766_160 |
0.0585911333 |
- |
- |
Serine/threonine-protein kinase SAPK1 [Gossypium arboreum] |
| 4 |
Hb_000088_230 |
0.0595619863 |
- |
- |
hypothetical protein POPTR_0013s11340g [Populus trichocarpa] |
| 5 |
Hb_001006_150 |
0.0615367626 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 6 |
Hb_001322_120 |
0.0624130066 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 7 |
Hb_000460_040 |
0.064589262 |
- |
- |
PREDICTED: protein phosphatase 2C 57 [Jatropha curcas] |
| 8 |
Hb_000445_270 |
0.0650260866 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_001338_120 |
0.0663987388 |
- |
- |
hypothetical chloroplast RF68 (chloroplast) [Ipomoea batatas] |
| 10 |
Hb_004306_090 |
0.066583253 |
- |
- |
PREDICTED: uncharacterized protein LOC105644392 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000359_050 |
0.0674686725 |
- |
- |
Oxygen-evolving enhancer protein 2, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_007477_060 |
0.0675964995 |
- |
- |
unknown [Medicago truncatula] |
| 13 |
Hb_000962_090 |
0.0698126553 |
- |
- |
omega-6 fatty acid desaturase [Hevea brasiliensis] |
| 14 |
Hb_002871_180 |
0.0731561391 |
- |
- |
photosystem II 10 kDa polypeptide, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001159_050 |
0.0764507942 |
- |
- |
PREDICTED: uncharacterized protein LOC105645952 [Jatropha curcas] |
| 16 |
Hb_003494_090 |
0.0785020423 |
transcription factor |
TF Family: MYB |
myb family transcription factor family protein [Populus trichocarpa] |
| 17 |
Hb_080922_010 |
0.0787124799 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 18 |
Hb_002272_210 |
0.0794929032 |
- |
- |
PREDICTED: UPF0603 protein At1g54780, chloroplastic [Jatropha curcas] |
| 19 |
Hb_005649_060 |
0.0806385045 |
- |
- |
PREDICTED: uncharacterized protein LOC105644161 [Jatropha curcas] |
| 20 |
Hb_001292_030 |
0.0817619254 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP26, chloroplastic [Jatropha curcas] |