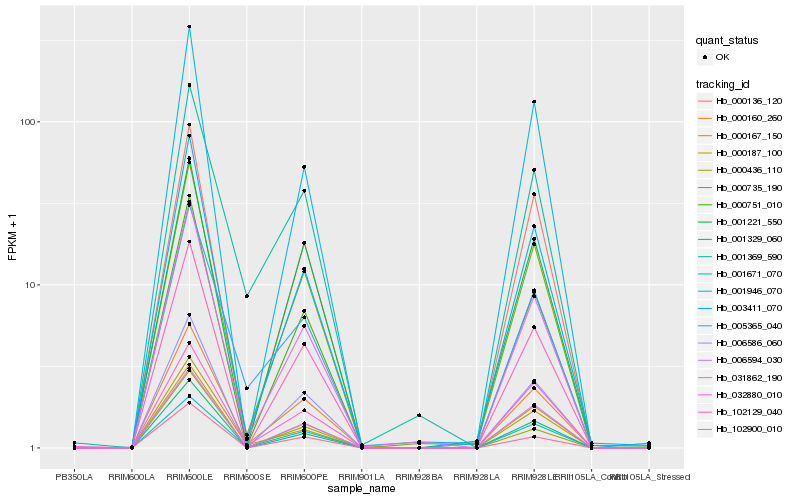
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000160_260 |
0.0 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 2 |
Hb_000735_190 |
0.0416570078 |
- |
- |
hypothetical protein POPTR_0006s19340g [Populus trichocarpa] |
| 3 |
Hb_001671_070 |
0.0520849733 |
- |
- |
kinase, putative [Ricinus communis] |
| 4 |
Hb_006586_060 |
0.0585650757 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 5 |
Hb_000751_010 |
0.059877905 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_102129_040 |
0.0608695423 |
- |
- |
PREDICTED: PTI1-like tyrosine-protein kinase 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_031862_190 |
0.0674324911 |
- |
- |
- |
| 8 |
Hb_003411_070 |
0.07536426 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 3.1 [Populus euphratica] |
| 9 |
Hb_000167_150 |
0.0771789338 |
- |
- |
hypothetical protein POPTR_0008s03590g [Populus trichocarpa] |
| 10 |
Hb_000136_120 |
0.0773062728 |
- |
- |
PREDICTED: probable strigolactone esterase D14 homolog [Jatropha curcas] |
| 11 |
Hb_005365_040 |
0.0784133003 |
- |
- |
hypothetical protein JCGZ_15100 [Jatropha curcas] |
| 12 |
Hb_001221_550 |
0.0792168382 |
- |
- |
Corticosteroid 11-beta-dehydrogenase, putative [Ricinus communis] |
| 13 |
Hb_102900_010 |
0.081390486 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001329_060 |
0.0819669657 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 15 |
Hb_001369_590 |
0.0824856119 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s14600g [Populus trichocarpa] |
| 16 |
Hb_000187_100 |
0.0840061424 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 17 |
Hb_006594_030 |
0.0893747066 |
- |
- |
- |
| 18 |
Hb_000436_110 |
0.090679865 |
- |
- |
PREDICTED: uncharacterized protein LOC105141197 [Populus euphratica] |
| 19 |
Hb_032880_010 |
0.0945801176 |
- |
- |
PREDICTED: beta-glucosidase 24-like [Jatropha curcas] |
| 20 |
Hb_001946_070 |
0.0947805282 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |