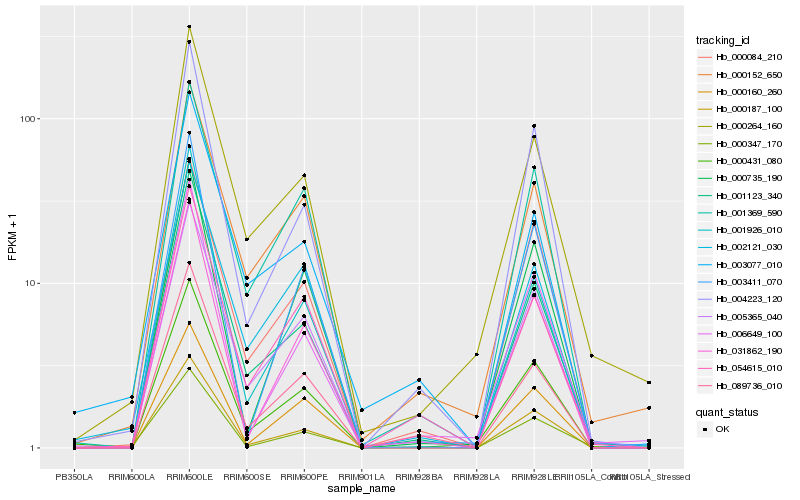
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000431_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632547 [Jatropha curcas] |
| 2 |
Hb_000187_100 |
0.0810327432 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 3 |
Hb_005365_040 |
0.0862855127 |
- |
- |
hypothetical protein JCGZ_15100 [Jatropha curcas] |
| 4 |
Hb_000264_160 |
0.0881659989 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX5-like [Jatropha curcas] |
| 5 |
Hb_001123_340 |
0.0935998737 |
- |
- |
PREDICTED: uncharacterized protein LOC105123089 isoform X1 [Populus euphratica] |
| 6 |
Hb_004223_120 |
0.0938669145 |
- |
- |
PREDICTED: protein SPA, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001369_590 |
0.0982774796 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s14600g [Populus trichocarpa] |
| 8 |
Hb_031862_190 |
0.0986857142 |
- |
- |
- |
| 9 |
Hb_006649_100 |
0.099284826 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 10 |
Hb_000152_650 |
0.1006217285 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 11 |
Hb_003411_070 |
0.1012467039 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 3.1 [Populus euphratica] |
| 12 |
Hb_000160_260 |
0.1034884982 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 13 |
Hb_089736_010 |
0.1092917695 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 7 [Populus euphratica] |
| 14 |
Hb_000735_190 |
0.1097070181 |
- |
- |
hypothetical protein POPTR_0006s19340g [Populus trichocarpa] |
| 15 |
Hb_002121_030 |
0.1107973478 |
- |
- |
Ribonuclease P protein subunit P38-related isoform 1 [Theobroma cacao] |
| 16 |
Hb_001926_010 |
0.1119405446 |
- |
- |
hypothetical protein Csa_2G383910 [Cucumis sativus] |
| 17 |
Hb_000347_170 |
0.1119472454 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 18 |
Hb_000084_210 |
0.1138919415 |
- |
- |
hypothetical protein POPTR_0016s08050g [Populus trichocarpa] |
| 19 |
Hb_054615_010 |
0.1173546236 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |
| 20 |
Hb_003077_010 |
0.1188622706 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit O, chloroplastic isoform X1 [Jatropha curcas] |