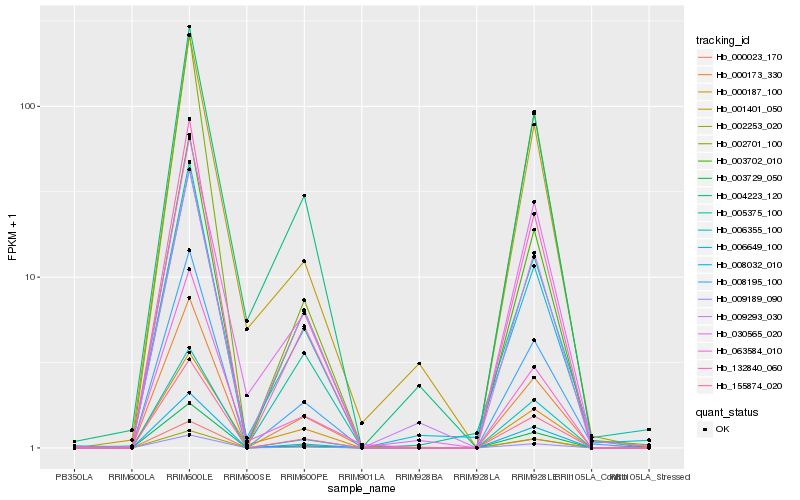
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001401_050 |
0.0 |
- |
- |
PREDICTED: chlorophyll a-b binding protein P4, chloroplastic [Jatropha curcas] |
| 2 |
Hb_004223_120 |
0.0801026144 |
- |
- |
PREDICTED: protein SPA, chloroplastic [Jatropha curcas] |
| 3 |
Hb_030565_020 |
0.0857096325 |
- |
- |
PREDICTED: protein ECERIFERUM 26 [Jatropha curcas] |
| 4 |
Hb_008032_010 |
0.0982193807 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 5 |
Hb_132840_060 |
0.0984483195 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH57-like [Jatropha curcas] |
| 6 |
Hb_006355_100 |
0.0988362925 |
- |
- |
hypothetical protein glysoja_002455 [Glycine soja] |
| 7 |
Hb_003729_050 |
0.0999039253 |
- |
- |
PREDICTED: vacuolar iron transporter 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_063584_010 |
0.1006901534 |
- |
- |
hypothetical protein JCGZ_10530 [Jatropha curcas] |
| 9 |
Hb_009189_090 |
0.1036680326 |
- |
- |
protein phosphatase-2c, putative [Ricinus communis] |
| 10 |
Hb_000023_170 |
0.1067967448 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2-like [Jatropha curcas] |
| 11 |
Hb_002701_100 |
0.107157597 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 12 |
Hb_006649_100 |
0.1080985696 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 13 |
Hb_155874_020 |
0.1088302646 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 14 |
Hb_003702_010 |
0.1103737188 |
- |
- |
PREDICTED: phospholipase A1-II 1-like [Jatropha curcas] |
| 15 |
Hb_008195_100 |
0.1113743522 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: protein YABBY 4 [Vitis vinifera] |
| 16 |
Hb_000187_100 |
0.1118168836 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 17 |
Hb_005375_100 |
0.1120136215 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH57-like [Jatropha curcas] |
| 18 |
Hb_000173_330 |
0.1151416208 |
- |
- |
PREDICTED: glutamine synthetase leaf isozyme, chloroplastic [Vitis vinifera] |
| 19 |
Hb_002253_020 |
0.1175208949 |
- |
- |
PREDICTED: callose synthase 5 [Jatropha curcas] |
| 20 |
Hb_009293_030 |
0.1192802615 |
- |
- |
PREDICTED: salicylate carboxymethyltransferase [Jatropha curcas] |