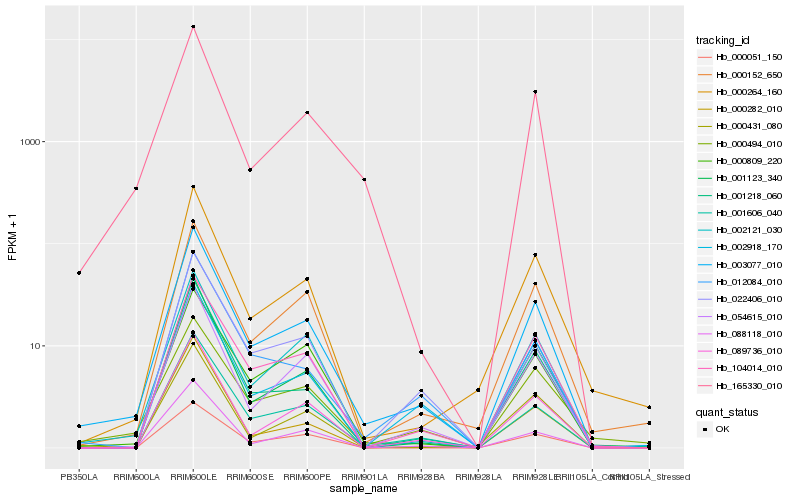
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003077_010 |
0.0 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit O, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_001606_040 |
0.0955230039 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 3 |
Hb_001123_340 |
0.0994935563 |
- |
- |
PREDICTED: uncharacterized protein LOC105123089 isoform X1 [Populus euphratica] |
| 4 |
Hb_104014_010 |
0.1047577348 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39020 isoform X2 [Jatropha curcas] |
| 5 |
Hb_089736_010 |
0.1055153372 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 7 [Populus euphratica] |
| 6 |
Hb_001218_060 |
0.1058564991 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002121_030 |
0.1068135933 |
- |
- |
Ribonuclease P protein subunit P38-related isoform 1 [Theobroma cacao] |
| 8 |
Hb_000152_650 |
0.1080646719 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 9 |
Hb_000264_160 |
0.1091159837 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX5-like [Jatropha curcas] |
| 10 |
Hb_054615_010 |
0.1104437697 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |
| 11 |
Hb_088118_010 |
0.1157971041 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 12 |
Hb_002918_170 |
0.1170273333 |
- |
- |
PREDICTED: MACPF domain-containing protein NSL1 [Jatropha curcas] |
| 13 |
Hb_012084_010 |
0.1186031248 |
- |
- |
cold regulated protein [Manihot esculenta] |
| 14 |
Hb_165330_010 |
0.1186426754 |
- |
- |
BnaUnng00840D [Brassica napus] |
| 15 |
Hb_000431_080 |
0.1188622706 |
- |
- |
PREDICTED: uncharacterized protein LOC105632547 [Jatropha curcas] |
| 16 |
Hb_000494_010 |
0.1228485795 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0014s04460g [Populus trichocarpa] |
| 17 |
Hb_000051_150 |
0.1228828869 |
- |
- |
hypothetical protein CICLE_v10003316mg [Citrus clementina] |
| 18 |
Hb_022406_010 |
0.1233487457 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 19 |
Hb_000282_010 |
0.1242083796 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000809_220 |
0.1281910869 |
- |
- |
PREDICTED: 3-ketodihydrosphingosine reductase [Jatropha curcas] |