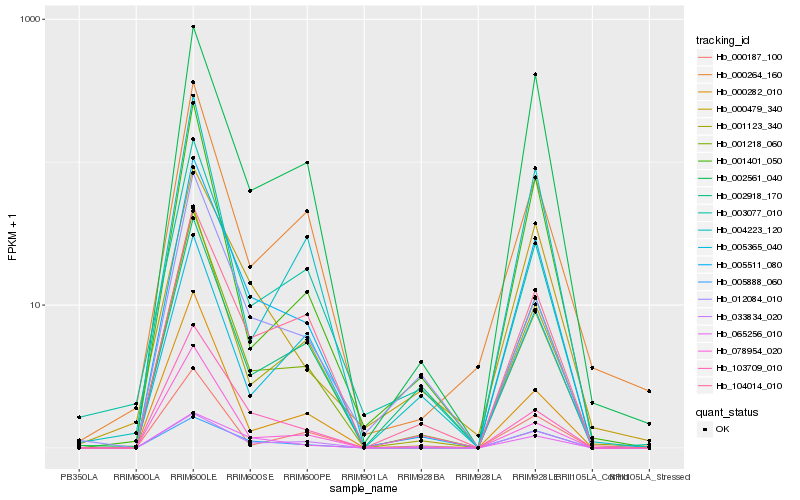
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005511_080 |
0.0 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 2 |
Hb_001218_060 |
0.0821992613 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_104014_010 |
0.1120361964 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39020 isoform X2 [Jatropha curcas] |
| 4 |
Hb_001123_340 |
0.1131453428 |
- |
- |
PREDICTED: uncharacterized protein LOC105123089 isoform X1 [Populus euphratica] |
| 5 |
Hb_002561_040 |
0.1184882623 |
- |
- |
Photosynthetic electron transfer C [Theobroma cacao] |
| 6 |
Hb_065256_010 |
0.1203039319 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 7 |
Hb_005888_060 |
0.1241418597 |
- |
- |
hypothetical protein PRUPE_ppa002432m2g, partial [Prunus persica] |
| 8 |
Hb_033834_020 |
0.1334983268 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000479_340 |
0.133973739 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 10 |
Hb_003077_010 |
0.1354351991 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit O, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_002918_170 |
0.1355910563 |
- |
- |
PREDICTED: MACPF domain-containing protein NSL1 [Jatropha curcas] |
| 12 |
Hb_078954_020 |
0.1357461835 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 13 |
Hb_000264_160 |
0.136494023 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX5-like [Jatropha curcas] |
| 14 |
Hb_000282_010 |
0.1408601912 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001401_050 |
0.1419688877 |
- |
- |
PREDICTED: chlorophyll a-b binding protein P4, chloroplastic [Jatropha curcas] |
| 16 |
Hb_005365_040 |
0.1424714879 |
- |
- |
hypothetical protein JCGZ_15100 [Jatropha curcas] |
| 17 |
Hb_004223_120 |
0.1444525367 |
- |
- |
PREDICTED: protein SPA, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000187_100 |
0.1454549642 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 19 |
Hb_012084_010 |
0.146073026 |
- |
- |
cold regulated protein [Manihot esculenta] |
| 20 |
Hb_103709_010 |
0.1462871006 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |