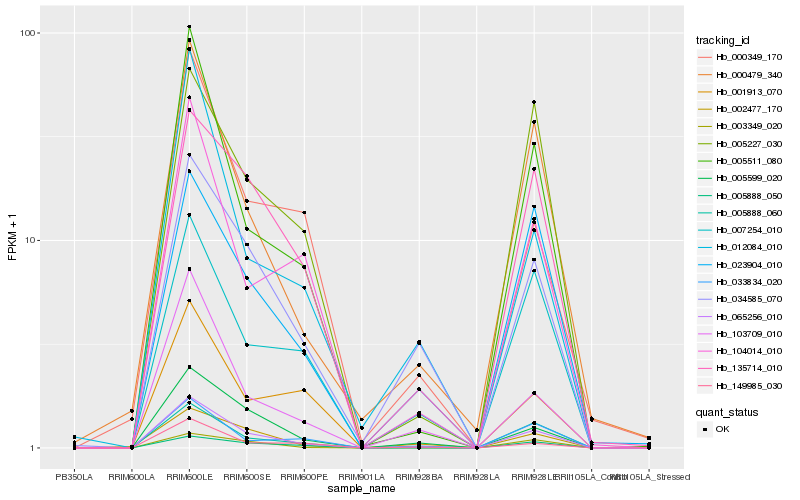
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_065256_010 |
0.0 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 2 |
Hb_005511_080 |
0.1203039319 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 3 |
Hb_005599_020 |
0.1231131152 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 4 |
Hb_034585_070 |
0.1297868808 |
- |
- |
Polyneuridine-aldehyde esterase precursor, putative [Ricinus communis] |
| 5 |
Hb_005888_060 |
0.1317101275 |
- |
- |
hypothetical protein PRUPE_ppa002432m2g, partial [Prunus persica] |
| 6 |
Hb_000479_340 |
0.1329405932 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 7 |
Hb_103709_010 |
0.1401989322 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 8 |
Hb_135714_010 |
0.1469924738 |
- |
- |
hypothetical protein POPTR_0017s09310g, partial [Populus trichocarpa] |
| 9 |
Hb_002477_170 |
0.148158032 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_007254_010 |
0.1493894515 |
- |
- |
hypothetical protein JCGZ_25214 [Jatropha curcas] |
| 11 |
Hb_104014_010 |
0.154733439 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39020 isoform X2 [Jatropha curcas] |
| 12 |
Hb_149985_030 |
0.1547884402 |
- |
- |
PREDICTED: zeatin O-glucosyltransferase-like [Jatropha curcas] |
| 13 |
Hb_005888_050 |
0.1557231522 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_033834_020 |
0.1563236363 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003349_020 |
0.1568141557 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Beta vulgaris subsp. vulgaris] |
| 16 |
Hb_023904_010 |
0.1598347731 |
- |
- |
- |
| 17 |
Hb_000349_170 |
0.1645484456 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 18 |
Hb_001913_070 |
0.1656052192 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |
| 19 |
Hb_005227_030 |
0.1698378607 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP28, chloroplastic [Jatropha curcas] |
| 20 |
Hb_012084_010 |
0.1717327892 |
- |
- |
cold regulated protein [Manihot esculenta] |