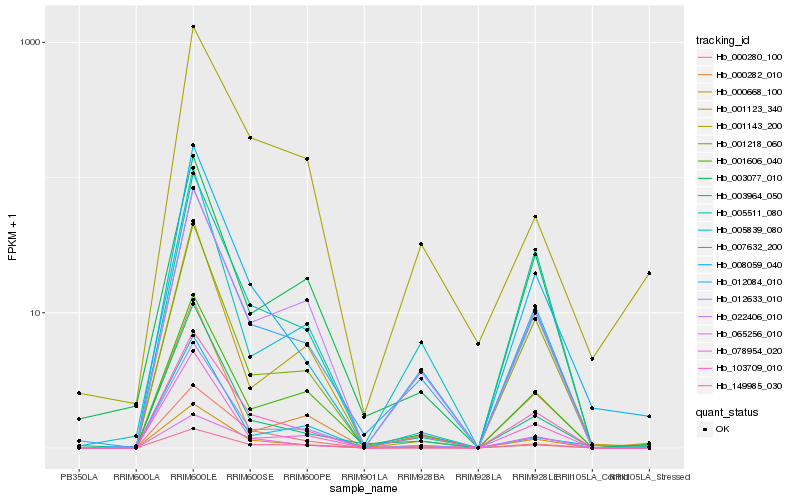
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008059_040 |
0.0 |
- |
- |
hypothetical protein POPTR_0019s12360g [Populus trichocarpa] |
| 2 |
Hb_103709_010 |
0.1085902829 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 3 |
Hb_012084_010 |
0.1122682178 |
- |
- |
cold regulated protein [Manihot esculenta] |
| 4 |
Hb_003964_050 |
0.1149185178 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor 25-like [Jatropha curcas] |
| 5 |
Hb_001218_060 |
0.1379972834 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_078954_020 |
0.1456879753 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 7 |
Hb_005839_080 |
0.157008644 |
- |
- |
Transcription factor bHLH61 isoform 1 [Theobroma cacao] |
| 8 |
Hb_012633_010 |
0.1571868114 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 49 [Jatropha curcas] |
| 9 |
Hb_000282_010 |
0.1594717902 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001606_040 |
0.1690533673 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 11 |
Hb_005511_080 |
0.1696098356 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 12 |
Hb_000280_100 |
0.1755491601 |
- |
- |
hypothetical protein JCGZ_13378 [Jatropha curcas] |
| 13 |
Hb_022406_010 |
0.1788375707 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 14 |
Hb_149985_030 |
0.1820788299 |
- |
- |
PREDICTED: zeatin O-glucosyltransferase-like [Jatropha curcas] |
| 15 |
Hb_001123_340 |
0.1840776405 |
- |
- |
PREDICTED: uncharacterized protein LOC105123089 isoform X1 [Populus euphratica] |
| 16 |
Hb_000668_100 |
0.1844936097 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 17 |
Hb_007632_200 |
0.1875071587 |
- |
- |
hypothetical protein L484_018221 [Morus notabilis] |
| 18 |
Hb_003077_010 |
0.1893871609 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit O, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_001143_200 |
0.1898067478 |
transcription factor |
TF Family: NAC |
NAC domain protein [Glycine max] |
| 20 |
Hb_065256_010 |
0.1922278715 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |