| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
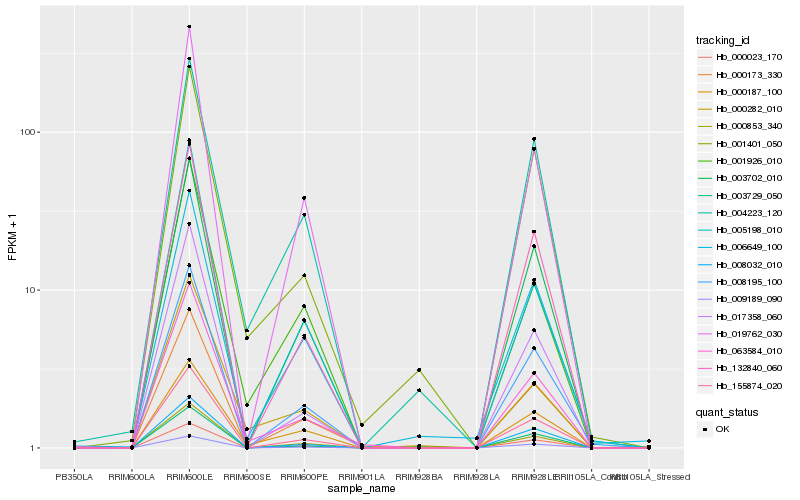
Hb_063584_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_10530 [Jatropha curcas] |
| 2 |
Hb_000853_340 |
0.0838442532 |
- |
- |
PREDICTED: kynurenine--oxoglutarate transaminase 1-like isoform X2 [Populus euphratica] |
| 3 |
Hb_155874_020 |
0.0839209718 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 4 |
Hb_019762_030 |
0.0902320778 |
- |
- |
hypothetical protein POPTR_0002s08720g [Populus trichocarpa] |
| 5 |
Hb_000173_330 |
0.0917172236 |
- |
- |
PREDICTED: glutamine synthetase leaf isozyme, chloroplastic [Vitis vinifera] |
| 6 |
Hb_132840_060 |
0.0934503248 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH57-like [Jatropha curcas] |
| 7 |
Hb_017358_060 |
0.0940855819 |
- |
- |
PREDICTED: uncharacterized protein LOC105641510 [Jatropha curcas] |
| 8 |
Hb_003702_010 |
0.0958682572 |
- |
- |
PREDICTED: phospholipase A1-II 1-like [Jatropha curcas] |
| 9 |
Hb_008195_100 |
0.0960311946 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: protein YABBY 4 [Vitis vinifera] |
| 10 |
Hb_001926_010 |
0.0967876574 |
- |
- |
hypothetical protein Csa_2G383910 [Cucumis sativus] |
| 11 |
Hb_000282_010 |
0.1000268939 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000187_100 |
0.1002349675 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 13 |
Hb_001401_050 |
0.1006901534 |
- |
- |
PREDICTED: chlorophyll a-b binding protein P4, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000023_170 |
0.103092027 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2-like [Jatropha curcas] |
| 15 |
Hb_008032_010 |
0.1051970384 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 16 |
Hb_003729_050 |
0.1064774089 |
- |
- |
PREDICTED: vacuolar iron transporter 1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_009189_090 |
0.1081768367 |
- |
- |
protein phosphatase-2c, putative [Ricinus communis] |
| 18 |
Hb_005198_010 |
0.1091357351 |
transcription factor |
TF Family: C2C2-YABBY |
PREDICTED: axial regulator YABBY 5 [Jatropha curcas] |
| 19 |
Hb_006649_100 |
0.1092549712 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 20 |
Hb_004223_120 |
0.1120534661 |
- |
- |
PREDICTED: protein SPA, chloroplastic [Jatropha curcas] |