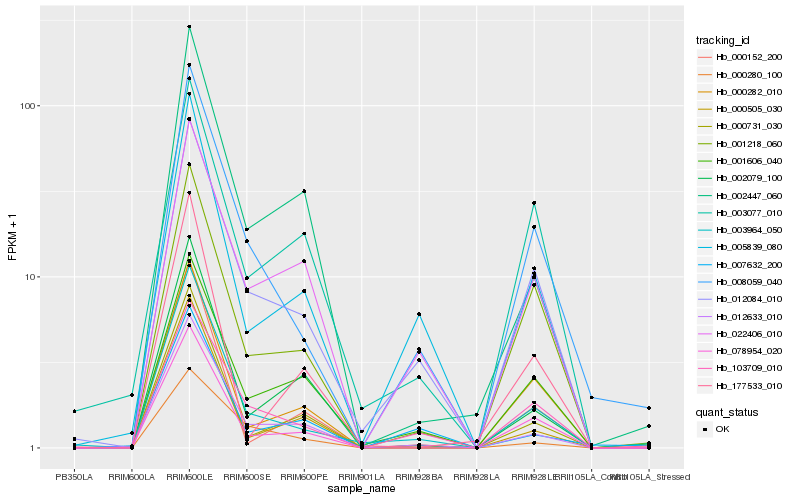
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003964_050 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor 25-like [Jatropha curcas] |
| 2 |
Hb_012084_010 |
0.1054234383 |
- |
- |
cold regulated protein [Manihot esculenta] |
| 3 |
Hb_008059_040 |
0.1149185178 |
- |
- |
hypothetical protein POPTR_0019s12360g [Populus trichocarpa] |
| 4 |
Hb_012633_010 |
0.1222621375 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 49 [Jatropha curcas] |
| 5 |
Hb_005839_080 |
0.1290582014 |
- |
- |
Transcription factor bHLH61 isoform 1 [Theobroma cacao] |
| 6 |
Hb_078954_020 |
0.1315862778 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 7 |
Hb_103709_010 |
0.1434151581 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 8 |
Hb_000282_010 |
0.1438560832 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001218_060 |
0.1497512498 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000505_030 |
0.1602991207 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002079_100 |
0.1635400549 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG5 [Jatropha curcas] |
| 12 |
Hb_000731_030 |
0.1640320327 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 13 |
Hb_001606_040 |
0.1669225729 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 14 |
Hb_000152_200 |
0.1670911405 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase ERL2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002447_060 |
0.1689158071 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_177533_010 |
0.176228047 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 17 |
Hb_000280_100 |
0.1762750985 |
- |
- |
hypothetical protein JCGZ_13378 [Jatropha curcas] |
| 18 |
Hb_007632_200 |
0.1766421449 |
- |
- |
hypothetical protein L484_018221 [Morus notabilis] |
| 19 |
Hb_003077_010 |
0.1776860833 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit O, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_022406_010 |
0.1786930822 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |