| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
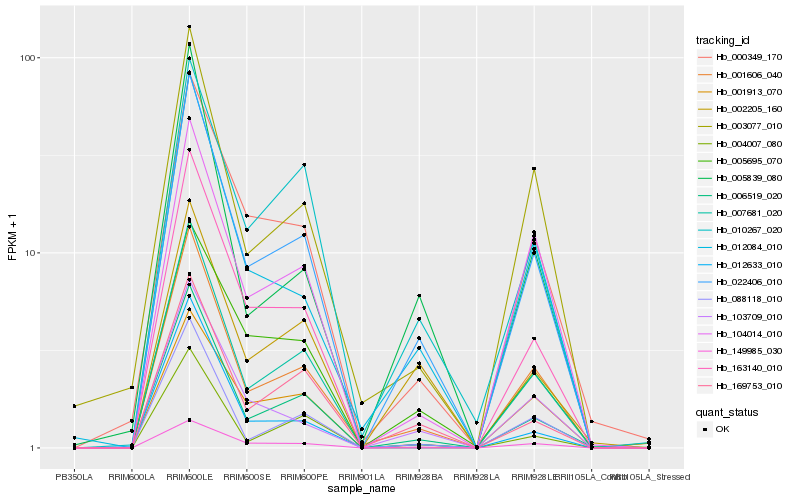
| 1 |
Hb_022406_010 |
0.0 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_149985_030 |
0.065025359 |
- |
- |
PREDICTED: zeatin O-glucosyltransferase-like [Jatropha curcas] |
| 3 |
Hb_001606_040 |
0.0663360711 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 4 |
Hb_006519_020 |
0.0695884021 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 5 |
Hb_012084_010 |
0.1018234715 |
- |
- |
cold regulated protein [Manihot esculenta] |
| 6 |
Hb_169753_010 |
0.105675275 |
- |
- |
hypothetical protein JCGZ_18439 [Jatropha curcas] |
| 7 |
Hb_005695_070 |
0.1093750646 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 8 |
Hb_000349_170 |
0.1094464508 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 9 |
Hb_103709_010 |
0.1114527956 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 10 |
Hb_010267_020 |
0.111565342 |
- |
- |
diphosphoinositol polyphosphate phosphohydrolase, putative [Ricinus communis] |
| 11 |
Hb_002205_160 |
0.1128073075 |
- |
- |
chalcone isomerase [Hibiscus cannabinus] |
| 12 |
Hb_007681_020 |
0.1134958735 |
- |
- |
transferase, putative [Ricinus communis] |
| 13 |
Hb_088118_010 |
0.1138167229 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 14 |
Hb_163140_010 |
0.1226345713 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 15 |
Hb_003077_010 |
0.1233487457 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit O, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_001913_070 |
0.1242938936 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |
| 17 |
Hb_104014_010 |
0.1242962433 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39020 isoform X2 [Jatropha curcas] |
| 18 |
Hb_004007_080 |
0.1244209807 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g49900 [Jatropha curcas] |
| 19 |
Hb_012633_010 |
0.1263993342 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 49 [Jatropha curcas] |
| 20 |
Hb_005839_080 |
0.1279999825 |
- |
- |
Transcription factor bHLH61 isoform 1 [Theobroma cacao] |