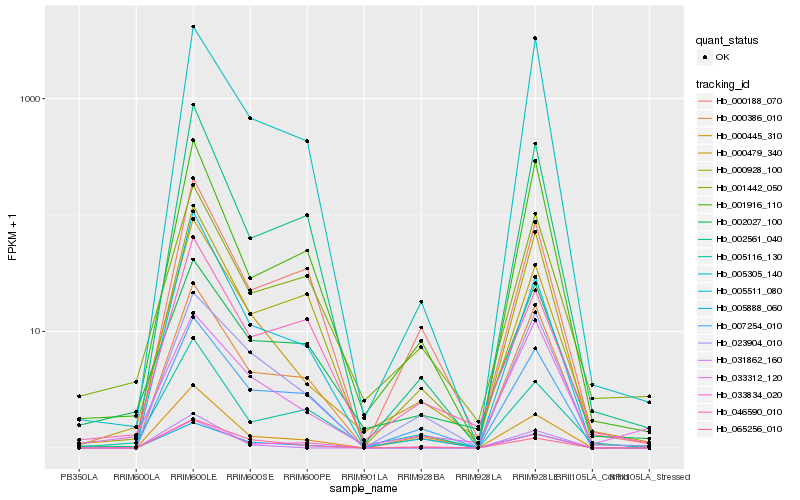
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000479_340 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 2 |
Hb_065256_010 |
0.1329405932 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 3 |
Hb_005888_060 |
0.1330249551 |
- |
- |
hypothetical protein PRUPE_ppa002432m2g, partial [Prunus persica] |
| 4 |
Hb_005511_080 |
0.133973739 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 5 |
Hb_023904_010 |
0.1491173403 |
- |
- |
- |
| 6 |
Hb_000386_010 |
0.1581247837 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002561_040 |
0.1583733283 |
- |
- |
Photosynthetic electron transfer C [Theobroma cacao] |
| 8 |
Hb_033834_020 |
0.1603845969 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000445_310 |
0.1637626757 |
- |
- |
hypothetical protein B456_011G218000 [Gossypium raimondii] |
| 10 |
Hb_007254_010 |
0.1642038548 |
- |
- |
hypothetical protein JCGZ_25214 [Jatropha curcas] |
| 11 |
Hb_031862_160 |
0.1645503639 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_001442_050 |
0.170759621 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit U, chloroplastic [Jatropha curcas] |
| 13 |
Hb_005116_130 |
0.1721234696 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa a, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000928_100 |
0.1722846006 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 15 |
Hb_001916_110 |
0.1726119578 |
- |
- |
PREDICTED: uncharacterized protein LOC105641510 [Jatropha curcas] |
| 16 |
Hb_005305_140 |
0.1735724352 |
- |
- |
unknown [Populus trichocarpa] |
| 17 |
Hb_002027_100 |
0.1743705753 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000188_070 |
0.1749205903 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 19 |
Hb_046590_010 |
0.1757473153 |
- |
- |
Cycloeucalenol cycloisomerase, putative [Ricinus communis] |
| 20 |
Hb_033312_120 |
0.1761391144 |
- |
- |
PREDICTED: psbQ-like protein 3, chloroplastic [Jatropha curcas] |