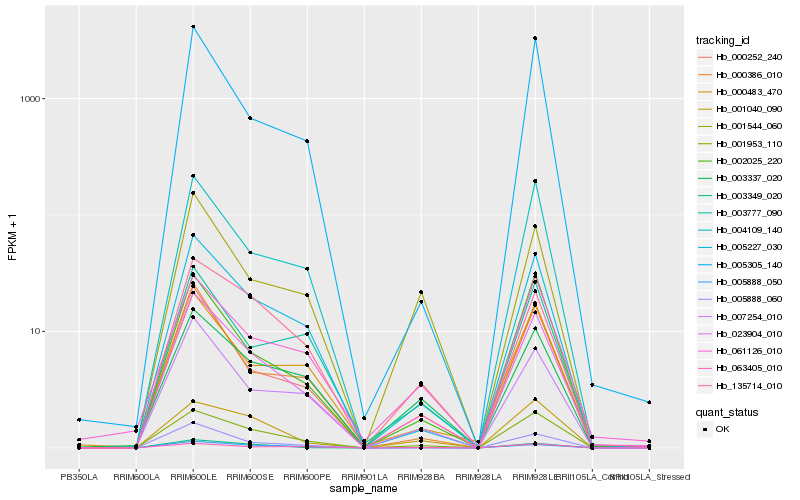
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023904_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_001953_110 |
0.083250435 |
- |
- |
PREDICTED: protein TAPETUM DETERMINANT 1-like [Jatropha curcas] |
| 3 |
Hb_005227_030 |
0.0972423406 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP28, chloroplastic [Jatropha curcas] |
| 4 |
Hb_007254_010 |
0.1044299745 |
- |
- |
hypothetical protein JCGZ_25214 [Jatropha curcas] |
| 5 |
Hb_000252_240 |
0.1050101297 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like isoform X2 [Gossypium raimondii] |
| 6 |
Hb_003337_020 |
0.1172953913 |
- |
- |
PREDICTED: uncharacterized protein LOC105116455 [Populus euphratica] |
| 7 |
Hb_000386_010 |
0.1181029249 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002025_220 |
0.1189308262 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_000483_470 |
0.1211578107 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_005305_140 |
0.1212236726 |
- |
- |
unknown [Populus trichocarpa] |
| 11 |
Hb_004109_140 |
0.1214680138 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_061126_010 |
0.1255079001 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 13 |
Hb_005888_050 |
0.127831549 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_135714_010 |
0.1285223902 |
- |
- |
hypothetical protein POPTR_0017s09310g, partial [Populus trichocarpa] |
| 15 |
Hb_003349_020 |
0.1287764021 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Beta vulgaris subsp. vulgaris] |
| 16 |
Hb_005888_060 |
0.1288548161 |
- |
- |
hypothetical protein PRUPE_ppa002432m2g, partial [Prunus persica] |
| 17 |
Hb_003777_090 |
0.1315174447 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 18 |
Hb_001544_060 |
0.131957611 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB4 isoform X2 [Jatropha curcas] |
| 19 |
Hb_063405_010 |
0.1369494827 |
- |
- |
PREDICTED: probable terpene synthase 3 [Jatropha curcas] |
| 20 |
Hb_001040_090 |
0.1389954508 |
- |
- |
PREDICTED: tetraspanin-19-like [Jatropha curcas] |