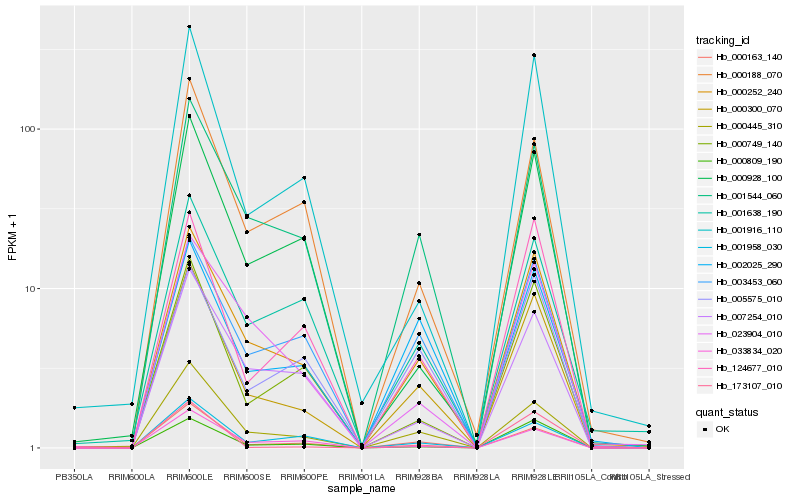
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_070 |
0.0 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 2 |
Hb_000252_240 |
0.081887045 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like isoform X2 [Gossypium raimondii] |
| 3 |
Hb_000445_310 |
0.0986956903 |
- |
- |
hypothetical protein B456_011G218000 [Gossypium raimondii] |
| 4 |
Hb_000163_140 |
0.1064464849 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 5 |
Hb_001544_060 |
0.1182104795 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB4 isoform X2 [Jatropha curcas] |
| 6 |
Hb_124677_010 |
0.1329207786 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002025_290 |
0.1368230993 |
transcription factor |
TF Family: G2-like |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000749_140 |
0.1392445333 |
- |
- |
PREDICTED: 2-succinylbenzoate--CoA ligase, chloroplastic/peroxisomal [Jatropha curcas] |
| 9 |
Hb_003453_060 |
0.1423843021 |
- |
- |
PREDICTED: nudix hydrolase 13, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000809_190 |
0.1425269056 |
- |
- |
PREDICTED: myosin-6-like [Jatropha curcas] |
| 11 |
Hb_001916_110 |
0.1459834493 |
- |
- |
PREDICTED: uncharacterized protein LOC105641510 [Jatropha curcas] |
| 12 |
Hb_005575_010 |
0.149008922 |
- |
- |
hypothetical protein POPTR_0004s02560g [Populus trichocarpa] |
| 13 |
Hb_001958_030 |
0.1499581008 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 14 |
Hb_000188_070 |
0.1569891441 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 15 |
Hb_033834_020 |
0.1592534725 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001638_190 |
0.1594412821 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_023904_010 |
0.1601324541 |
- |
- |
- |
| 18 |
Hb_007254_010 |
0.1602345141 |
- |
- |
hypothetical protein JCGZ_25214 [Jatropha curcas] |
| 19 |
Hb_000928_100 |
0.1635899222 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 20 |
Hb_173107_010 |
0.1637680208 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |