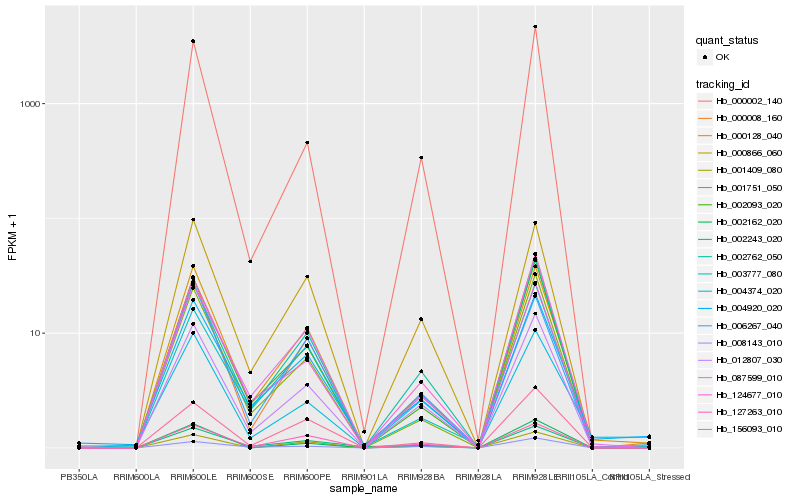
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012807_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628042 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002762_050 |
0.0717904906 |
- |
- |
PREDICTED: uncharacterized protein LOC105640359 [Jatropha curcas] |
| 3 |
Hb_124677_010 |
0.0840347378 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000002_140 |
0.0854866412 |
- |
- |
hypothetical protein JCGZ_02318 [Jatropha curcas] |
| 5 |
Hb_000008_160 |
0.0867853899 |
- |
- |
carboxy-lyase, putative [Ricinus communis] |
| 6 |
Hb_000866_060 |
0.0881580809 |
- |
- |
putative glycerol-3-phosphate acyltransferase [Vernicia fordii] |
| 7 |
Hb_000128_040 |
0.0925147295 |
- |
- |
PREDICTED: uncharacterized protein LOC105641228 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002093_020 |
0.0929596998 |
- |
- |
hypothetical protein POPTR_0008s02960g [Populus trichocarpa] |
| 9 |
Hb_004920_020 |
0.0970983238 |
- |
- |
hypothetical protein POPTR_0014s11700g [Populus trichocarpa] |
| 10 |
Hb_001751_050 |
0.0990134537 |
- |
- |
PREDICTED: phytol kinase 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001409_080 |
0.0990941224 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X1 [Populus euphratica] |
| 12 |
Hb_008143_010 |
0.099964033 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 13 |
Hb_004374_020 |
0.112094269 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog MMK2-like [Jatropha curcas] |
| 14 |
Hb_002162_020 |
0.112251279 |
- |
- |
hypothetical protein JCGZ_21655 [Jatropha curcas] |
| 15 |
Hb_087599_010 |
0.1144791237 |
- |
- |
PREDICTED: tropinone reductase homolog [Solanum tuberosum] |
| 16 |
Hb_127263_010 |
0.1198811436 |
- |
- |
hypothetical protein JCGZ_02828 [Jatropha curcas] |
| 17 |
Hb_002243_020 |
0.1236458653 |
- |
- |
PREDICTED: uncharacterized protein LOC105642097 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003777_080 |
0.1238361881 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Populus euphratica] |
| 19 |
Hb_156093_010 |
0.1241393028 |
- |
- |
hypothetical protein RCOM_0504250 [Ricinus communis] |
| 20 |
Hb_006267_040 |
0.1260144425 |
- |
- |
PREDICTED: ABC transporter G family member 32 isoform X1 [Jatropha curcas] |