| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
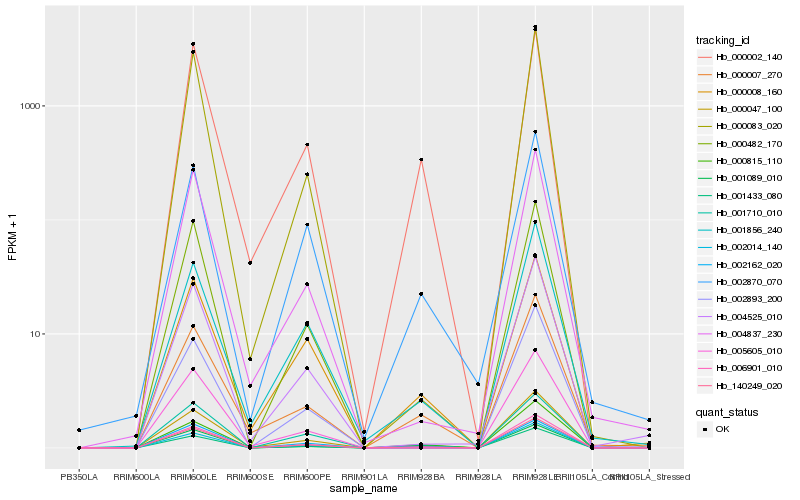
Hb_140249_020 |
0.0 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein CMB1-like [Jatropha curcas] |
| 2 |
Hb_001433_080 |
0.0316913728 |
- |
- |
Pectinesterase precursor, putative [Ricinus communis] |
| 3 |
Hb_000047_100 |
0.0502843978 |
transcription factor |
TF Family: GRF |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002162_020 |
0.0749520262 |
- |
- |
hypothetical protein JCGZ_21655 [Jatropha curcas] |
| 5 |
Hb_000815_110 |
0.0850488205 |
- |
- |
RecName: Full=Casparian strip membrane protein 4; Short=RcCASP4 [Ricinus communis] |
| 6 |
Hb_000002_140 |
0.0865257512 |
- |
- |
hypothetical protein JCGZ_02318 [Jatropha curcas] |
| 7 |
Hb_000007_270 |
0.091861528 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006901_010 |
0.0935806319 |
- |
- |
hypothetical protein JCGZ_20722 [Jatropha curcas] |
| 9 |
Hb_001089_010 |
0.093994309 |
- |
- |
PREDICTED: GDSL esterase/lipase 5-like [Jatropha curcas] |
| 10 |
Hb_001710_010 |
0.0941798791 |
- |
- |
Uncharacterized protein TCM_001832 [Theobroma cacao] |
| 11 |
Hb_002014_140 |
0.0967157535 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 12 |
Hb_000482_170 |
0.0968212489 |
transcription factor |
TF Family: C2C2-GATA |
GATA nirate-inducible carbon-metabolism involved protein [Populus nigra x Populus x canadensis] |
| 13 |
Hb_000083_020 |
0.0971448944 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 14 |
Hb_000008_160 |
0.0977591985 |
- |
- |
carboxy-lyase, putative [Ricinus communis] |
| 15 |
Hb_004525_010 |
0.100008281 |
- |
- |
late embryogenesis abundant, putative [Ricinus communis] |
| 16 |
Hb_004837_230 |
0.1020821776 |
- |
- |
unknown [Populus trichocarpa] |
| 17 |
Hb_002893_200 |
0.1023543708 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_002870_070 |
0.102517065 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 19 |
Hb_005605_010 |
0.1031559474 |
- |
- |
hypothetical protein POPTR_0019s13220g [Populus trichocarpa] |
| 20 |
Hb_001856_240 |
0.1037634539 |
transcription factor |
TF Family: TCP |
Plastid transcription factor 1, putative isoform 1 [Theobroma cacao] |