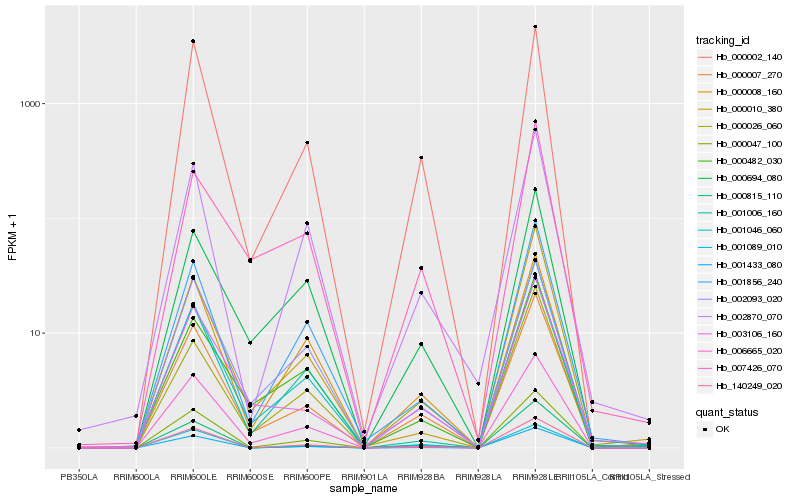
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000007_270 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000047_100 |
0.0760564457 |
transcription factor |
TF Family: GRF |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003106_160 |
0.0794085043 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g67520 [Jatropha curcas] |
| 4 |
Hb_140249_020 |
0.091861528 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein CMB1-like [Jatropha curcas] |
| 5 |
Hb_000002_140 |
0.0951418121 |
- |
- |
hypothetical protein JCGZ_02318 [Jatropha curcas] |
| 6 |
Hb_000815_110 |
0.0955858899 |
- |
- |
RecName: Full=Casparian strip membrane protein 4; Short=RcCASP4 [Ricinus communis] |
| 7 |
Hb_001433_080 |
0.1002246009 |
- |
- |
Pectinesterase precursor, putative [Ricinus communis] |
| 8 |
Hb_001856_240 |
0.1012686682 |
transcription factor |
TF Family: TCP |
Plastid transcription factor 1, putative isoform 1 [Theobroma cacao] |
| 9 |
Hb_000008_160 |
0.1067119252 |
- |
- |
carboxy-lyase, putative [Ricinus communis] |
| 10 |
Hb_000026_060 |
0.1131147352 |
- |
- |
beta-1,3-glucanase [Manihot esculenta] |
| 11 |
Hb_000482_030 |
0.1132538925 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER-like isoform X1 [Populus euphratica] |
| 12 |
Hb_002093_020 |
0.1139301661 |
- |
- |
hypothetical protein POPTR_0008s02960g [Populus trichocarpa] |
| 13 |
Hb_000694_080 |
0.1145026909 |
- |
- |
phosphoribulose kinase, putative [Ricinus communis] |
| 14 |
Hb_000010_380 |
0.1163230766 |
- |
- |
hypothetical protein CICLE_v10020897mg [Citrus clementina] |
| 15 |
Hb_006665_020 |
0.1164099176 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 16 |
Hb_002870_070 |
0.116750275 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 17 |
Hb_007426_070 |
0.1185694291 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 18 |
Hb_001006_160 |
0.1191529108 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_001089_010 |
0.1198696103 |
- |
- |
PREDICTED: GDSL esterase/lipase 5-like [Jatropha curcas] |
| 20 |
Hb_001046_060 |
0.12071826 |
- |
- |
PREDICTED: UDP-glucose flavonoid 3-O-glucosyltransferase 7-like [Fragaria vesca subsp. vesca] |